Restriction of HIV-1 and other retroviruses by TRIM5
- PMID: 31312031
- PMCID: PMC6858284
- DOI: 10.1038/s41579-019-0225-2
Restriction of HIV-1 and other retroviruses by TRIM5
Abstract
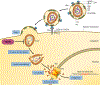
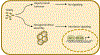
Mammalian cells express a variety of innate immune proteins - known as restriction factors - which defend against invading retroviruses such as HIV-1. Two members of the tripartite motif protein family - TRIM5α and TRIMCyp - were identified in 2004 as restriction factors that recognize and inactivate the capsid shell that surrounds and protects the incoming retroviral core. Research on these TRIM5 proteins has uncovered a novel mode of non-self recognition that protects against cross-species transmission of retroviruses. Our developing understanding of the mechanism of TRIM5 restriction underscores the concept that core uncoating and reverse transcription of the viral genome are coordinated processes rather than discrete steps of the post-entry pathway of retrovirus replication. In this Review, we provide an overview of the current state of knowledge of the molecular mechanism of TRIM5-mediated restriction, highlight recent advances and discuss implications for the development of capsid-targeted antiviral therapeutics.
Figures



References
-
- Shibata R, Sakai H, Kawamura M, Tokunaga K & Adachi A Early replication block of human immunodeficiency virus type 1 in monkey cells. J Gen Virol 76, 2723–2730 (1995). - PubMed
-
- Stremlau M et al. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427, 848–853 (2004). - PubMed
-
- Sayah DM, Sokolskaja E, Berthoux L & Luban J Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430, 569–573 (2004). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

