A Simulation Study Comparing Different Statistical Approaches for the Identification of Predictive Biomarkers
- PMID: 31312252
- PMCID: PMC6595324
- DOI: 10.1155/2019/7037230
A Simulation Study Comparing Different Statistical Approaches for the Identification of Predictive Biomarkers
Abstract
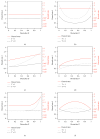
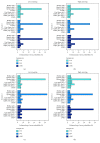
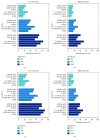
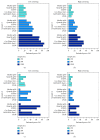
Identification of relevant biomarkers that are associated with a treatment effect is one requirement for adequate treatment stratification and consequently to improve health care by administering the best available treatment to an individual patient. Various statistical approaches were proposed that allow assessing the interaction between a continuous covariate and treatment. Nevertheless, categorization of a continuous covariate, e.g., by splitting the data at the observed median value, appears to be very prevalent in practice. In this article, we present a simulation study considering data as observed in a randomized clinical trial with a time-to-event outcome performed to compare properties of such approaches, namely, Cox regression with linear interaction, Multivariable Fractional Polynomials for Interaction (MFPI), Local Partial-Likelihood Bootstrap (LPLB), and the Subpopulation Treatment Effect Pattern Plot (STEPP) method, and of strategies based on categorization of continuous covariates (splitting the covariate at the median, splitting at quartiles, and using an "optimal" split by maximizing a corresponding test statistic). In different scenarios with no interactions, linear interactions or nonlinear interactions, type I error probability and the power for detection of a true covariate-treatment interaction were estimated. The Cox regression approach was more efficient than the other methods for scenarios with monotonous interactions, especially when the number of observed events was small to moderate. When patterns of the biomarker-treatment interaction effect were more complex, MFPI and LPLB performed well compared to the other approaches. Categorization of data generally led to a loss of power, but for very complex patterns, splitting the data into multiple categories might help to explore the nature of the interaction effect. Consequently, we recommend application of statistical methods developed for assessment of interactions between continuous biomarkers and treatment instead of arbitrary or data-driven categorization of continuous covariates.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

