A Multimodality Approach to Assessing Factor I Genetic Variants in Atypical Hemolytic Uremic Syndrome
- PMID: 31312772
- PMCID: PMC6609824
- DOI: 10.1016/j.ekir.2019.04.003
A Multimodality Approach to Assessing Factor I Genetic Variants in Atypical Hemolytic Uremic Syndrome
Figures
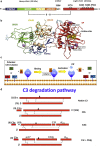
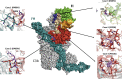



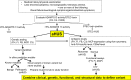
References
-
- Goodship T.H., Cook H.T., Fakhouri F. Atypical hemolytic uremic syndrome and C3 glomerulopathy: conclusions from a “Kidney Disease: Improving Global Outcomes“ (KDIGO) Controversies Conference. Kidney Int. 2017;91:539–551. - PubMed
-
- Kavanagh D., Richards A., Noris M. Characterization of mutations in complement factor I (CFI) associated with hemolytic uremic syndrome. Mol Immunol. 2007;45:95–105. - PubMed
-
- Le Quintrec M., Zuber J., Moulin B. Complement genes strongly predict recurrence and graft outcome in adult renal transplant recipients with atypical hemolytic and uremic syndrome. Am J Transplantation. 2013;13:663–675. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

