A gene regulatory network explains RET-EDNRB epistasis in Hirschsprung disease
- PMID: 31313802
- PMCID: PMC7275776
- DOI: 10.1093/hmg/ddz149
A gene regulatory network explains RET-EDNRB epistasis in Hirschsprung disease
Abstract
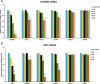
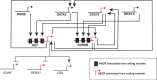
Disruptions in gene regulatory networks (GRNs), driven by multiple deleterious variants, potentially underlie complex traits and diseases. Hirschsprung disease (HSCR), a multifactorial disorder of enteric nervous system (ENS) development, is associated with at least 24 genes and seven chromosomal loci, with RET and EDNRB as its major genes. We previously demonstrated that RET transcription in the ENS is controlled by an extensive GRN involving the transcription factors (TFs) RARB, GATA2 and SOX10 and other HSCR genes. We now demonstrate, using human and mouse cellular and animal models, that EDNRB is transcriptionally regulated in the ENS by GATA2, SOX10 and NKX2.5 TFs. Significantly, RET and EDNRB expression is regulated by their shared use of GATA2 and SOX10, and in turn, these TFs are controlled by EDNRB and RET in a dose-dependent manner. This study expands the ENS development GRN to include both RET and EDNRB, uncovers the mechanistic basis for RET-EDNRB epistasis and emphasizes how functionally different genes associated with a complex disorder can be united through a common GRN.
© The Author(s) 2019. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures
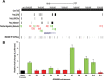
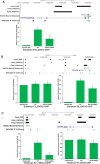
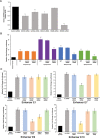
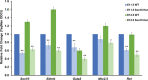
References
-
- Chakravarti A. and Turner T.N. (2016) Revealing rate-limiting steps in complex disease biology: the crucial importance of studying rare, extreme-phenotype families. Bioessays, 38, 578–586. - PubMed
-
- Davidson E.H. (2006) The regulatory genome: gene regulatory networks in development and evolution. Academic Press, Cambridge, MA, USA.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

