Shift in the paradigm towards next-generation microbiology
- PMID: 31314103
- PMCID: PMC6759065
- DOI: 10.1093/femsle/fnz159
Shift in the paradigm towards next-generation microbiology
Abstract
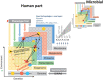
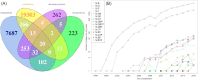
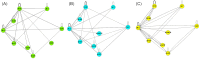
In this work, the position of contemporary microbiology is considered from the perspective of scientific success, and a list of historical points and lessons learned from the fields of medical microbiology, microbial ecology and systems biology is presented. In addition, patterns in the development of top-down research topics that emerged over time as well as overlapping ideas and personnel, which are the first signs of trans-domain research activities in the fields of metagenomics, metaproteomics, metatranscriptomics and metabolomics, are explored through analysis of the publication networks of 28 654 papers using the computer programme Pajek. The current state of affairs is defined, and the need for meta-analyses to leverage publication biases in the field of microbiology is put forward as a very important emerging field of microbiology, especially since microbiology is progressively dealing with multi-scale systems. Consequently, the need for cross-fertilisation with other fields/disciplines instead of 'more microbiology' is needed to advance the field of microbiology as such. The reader is directed to consider how novel technologies, the introduction of big data approaches and artificial intelligence have transformed microbiology into a multi-scale field and initiated a shift away from its history of mostly manual work and towards a largely technology-, data- and statistics-driven discipline that is often coupled with automation and modelling.
Keywords: bibliometric analysis; bioinformatics; biology; meta-analyses; metabolomics; metagenomics; metaproteomics; metatranscriptomics; publication bias; statistics; top-down.
© FEMS 2019.
Figures

Similar articles
-
The need for an integrated multi-OMICs approach in microbiome science in the food system.Compr Rev Food Sci Food Saf. 2023 Mar;22(2):1082-1103. doi: 10.1111/1541-4337.13103. Epub 2023 Jan 12. Compr Rev Food Sci Food Saf. 2023. PMID: 36636774
-
Molecular eco-systems biology: towards an understanding of community function.Nat Rev Microbiol. 2008 Sep;6(9):693-9. doi: 10.1038/nrmicro1935. Nat Rev Microbiol. 2008. PMID: 18587409 Review.
-
Predictive microbiology through the last century. From paper to Excel and towards AI.Adv Food Nutr Res. 2025;113:1-63. doi: 10.1016/bs.afnr.2024.09.012. Epub 2024 Oct 22. Adv Food Nutr Res. 2025. PMID: 40023558 Review.
-
Next generation microbiological risk assessment meta-omics: The next need for integration.Int J Food Microbiol. 2018 Dec 20;287:10-17. doi: 10.1016/j.ijfoodmicro.2017.11.008. Epub 2017 Nov 13. Int J Food Microbiol. 2018. PMID: 29157743
-
Prospects for multi-omics in the microbial ecology of water engineering.Water Res. 2021 Oct 15;205:117608. doi: 10.1016/j.watres.2021.117608. Epub 2021 Aug 27. Water Res. 2021. PMID: 34555741 Review.
Cited by
-
Lignin intermediates lead to phenyl acid formation and microbial community shifts in meso- and thermophilic batch reactors.Biotechnol Biofuels. 2021 Jan 20;14(1):27. doi: 10.1186/s13068-020-01855-0. Biotechnol Biofuels. 2021. PMID: 33472684 Free PMC article.
-
Fluorescent optotracers for bacterial and biofilm detection and diagnostics.Sci Technol Adv Mater. 2023 Sep 5;24(1):2246867. doi: 10.1080/14686996.2023.2246867. eCollection 2023. Sci Technol Adv Mater. 2023. PMID: 37680974 Free PMC article. Review.
-
A call to reignite the revolutionary spirit of scientific discovery.J Transl Med. 2025 Jun 24;23(1):699. doi: 10.1186/s12967-025-06669-y. J Transl Med. 2025. PMID: 40555985 Free PMC article. No abstract available.
-
An Early Stage Researcher's Primer on Systems Medicine Terminology.Netw Syst Med. 2021 Feb 25;4(1):2-50. doi: 10.1089/nsm.2020.0003. eCollection 2021 Feb. Netw Syst Med. 2021. PMID: 33659919 Free PMC article. Review.
-
SCIP: a self-paced, community-based summer coding program creates community and increases coding confidence, lessons learned from the pandemic.bioRxiv [Preprint]. 2024 Jul 4:2022.12.27.521952. doi: 10.1101/2022.12.27.521952. bioRxiv. 2024. PMID: 36597523 Free PMC article. Preprint.
References
-
- Allen B, Stacey BC, Bar-Yam Y. Multiscale information theory and the marginal utility of information. Entropy. 2017;19:273.
-
- Arrowsmith J. Phase II failures: 2008–2010. Nat Rev Drug Discov. 2011;10:328. - PubMed
-
- Asher M. Reliability of ‘new drug target’ claims called into question. Nat Rev Drug Discov. 2011;10:643. - PubMed
-
- Batagelj V. WoS2Pajek. Manual for Version 1.4. July2016, http://vladowiki.fmf.uni-lj.si/doku.php?id=pajek:wos2pajek (15 February 2018, date last accessed).

