Recombinant Hepatitis E Viruses Harboring Tags in the ORF1 Protein
- PMID: 31315997
- PMCID: PMC6744232
- DOI: 10.1128/JVI.00459-19
Recombinant Hepatitis E Viruses Harboring Tags in the ORF1 Protein
Abstract
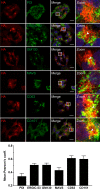
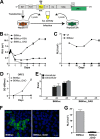
Hepatitis E virus (HEV) is one of the most common causes of acute hepatitis and jaundice in the world. Current understanding of the molecular virology and pathogenesis of hepatitis E is incomplete, due particularly to the limited availability of functional tools. Here, we report the development of tagged HEV genomes as a novel tool to investigate the viral life cycle. A selectable subgenomic HEV replicon was subjected to random 15-nucleotide sequence insertion using transposon-based technology. Viable insertions in the open reading frame 1 (ORF1) protein were selected in a hepatoblastoma cell line. Functional insertion sites were identified downstream of the methyltransferase domain, in the hypervariable region (HVR), and between the helicase and RNA-dependent RNA polymerase domains. HEV genomes harboring a hemagglutinin (HA) epitope tag or a small luciferase (NanoLuc) in the HVR were found to be fully functional and to allow the production of infectious virus. NanoLuc allowed quantitative monitoring of HEV infection and replication by luciferase assay. The use of HA-tagged replicons and full-length genomes allowed localization of putative sites of HEV RNA replication by the simultaneous detection of viral RNA by fluorescence in situ hybridization and of ORF1 protein by immunofluorescence. Candidate HEV replication complexes were found in cytoplasmic dot-like structures which partially overlapped ORF2 and ORF3 proteins as well as exosomal markers. Hence, tagged HEV genomes yield new insights into the viral life cycle and should allow further investigation of the structure and composition of the viral replication complex.IMPORTANCE Hepatitis E virus (HEV) infection is an important cause of acute hepatitis and may lead to chronic infection in immunocompromised patients. Knowledge of the viral life cycle is incomplete due to the limited availability of functional tools. In particular, low levels of expression of the ORF1 protein or limited sensitivity of currently available antibodies or both limit our understanding of the viral replicase. Here, we report the successful establishment of subgenomic HEV replicons and full-length genomes harboring an epitope tag or a functional reporter in the ORF1 protein. These novel tools should allow further characterization of the HEV replication complex and to improve our understanding of the viral life cycle.
Keywords: HEV; open reading frame 1 protein; positive-strand RNA virus; replicase; replication complex; replicon; transposon; viral life cycle.
Copyright © 2019 Szkolnicka et al.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

