The longevity-promoting factor, TCER-1, widely represses stress resistance and innate immunity
- PMID: 31316054
- PMCID: PMC6637209
- DOI: 10.1038/s41467-019-10759-z
The longevity-promoting factor, TCER-1, widely represses stress resistance and innate immunity
Abstract
Stress resistance and longevity are positively correlated but emerging evidence indicates that they are physiologically distinct. Identifying factors with distinctive roles in these processes is challenging because pro-longevity genes often enhance stress resistance. We demonstrate that TCER-1, the Caenorhabditis elegans homolog of human transcription elongation and splicing factor, TCERG1, has opposite effects on lifespan and stress resistance. We previously showed that tcer-1 promotes longevity in germline-less C. elegans and reproductive fitness in wild-type animals. Surprisingly, tcer-1 mutants exhibit exceptional resistance against multiple stressors, including infection by human opportunistic pathogens, whereas, TCER-1 overexpression confers immuno-susceptibility. TCER-1 inhibits immunity only during fertile stages of life. Elevating its levels ameliorates the fertility loss caused by infection, suggesting that TCER-1 represses immunity to augment fecundity. TCER-1 acts through repression of PMK-1 as well as PMK-1-independent factors critical for innate immunity. Our data establish key roles for TCER-1 in coordinating immunity, longevity and fertility, and reveal mechanisms that distinguish length of life from functional aspects of aging.
Conflict of interest statement
The authors declare no competing interests.
Figures



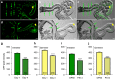
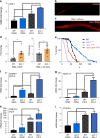

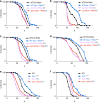
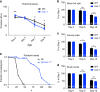
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

