SREBP1-dependent de novo fatty acid synthesis gene expression is elevated in malignant melanoma and represents a cellular survival trait
- PMID: 31316083
- PMCID: PMC6637239
- DOI: 10.1038/s41598-019-46594-x
SREBP1-dependent de novo fatty acid synthesis gene expression is elevated in malignant melanoma and represents a cellular survival trait
Abstract
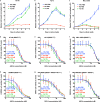
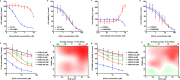
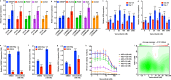
de novo fatty acid biosynthesis (DNFA) is a hallmark adaptation of many cancers that supports survival, proliferation, and metastasis. Here we elucidate previously unexplored aspects of transcription regulation and clinical relevance of DNFA in cancers. We show that elevated expression of DNFA genes is characteristic of many tumor types and correlates with poor prognosis, especially in melanomas. Elevated DNFA gene expression depends on the SREBP1 transcription factor in multiple melanoma cell lines. SREBP1 predominantly binds to the transcription start sites of DNFA genes, regulating their expression by recruiting RNA polymerase II to promoters for productive transcription elongation. We find that SREBP1-regulated DNFA represents a survival trait in melanoma cells, regardless of proliferative state and oncogenic mutation status. Indeed, malignant melanoma cells exhibit elevated DNFA gene expression after the BRAF/MEK signaling pathway is blocked (e.g. by BRAF inhibitors), and DNFA expression remains higher in melanoma cells resistant to vemurafenib treatment than in untreated cells. Accordingly, DNFA pathway inhibition, whether by direct targeting of SREBP1 with antisense oligonucleotides, or through combinatorial effects of multiple DNFA enzyme inhibitors, exerts potent cytotoxic effects on both BRAFi-sensitive and -resistant melanoma cells. Altogether, these results implicate SREBP1 and DNFA enzymes as enticing therapeutic targets in melanomas.
Conflict of interest statement
The authors declare no competing interests.
Figures