Sperm-borne miR-216b modulates cell proliferation during early embryo development via K-RAS
- PMID: 31316130
- PMCID: PMC6637201
- DOI: 10.1038/s41598-019-46775-8
Sperm-borne miR-216b modulates cell proliferation during early embryo development via K-RAS
Abstract
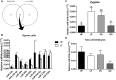
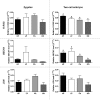
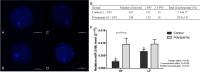
Semen fertilizing potential is dependent upon the morphological, functional and molecular attributes of sperm. Sperm microRNAs (miRNAs) were recently shown to hold promise regarding their association with different fertility phenotypes. However, their role in fertility regulation remains to be determined. We postulated that sperm miRNAs might regulate early embryonic development. From this perspective, sperm quality and 380 sperm miRNAs were investigated in frozen-thawed semen from high (HF; 54.3 ± 1.0% pregnancy rate) and low (LF; 41.5 ± 2.3%) fertility bulls. Out of nine miRNAs that showed different levels in sperm cells, miR-216b was present at lower levels in HF sperm cells and zygotes. Among miR-216b target genes (K-RAS, BECN1 and JUN), K-RAS, related to cell proliferation, revealed a higher level in HF two-cell embryos. First cleavage rate, blastocyst cell number and division number were also higher in HF. In addition, by using a model based on polyspermy embryos, we demonstrated an increase in miR-216b levels in zygotes associated with sperm cell entry. Our results shed light on a possible mechanism of paternal contribution involving sperm-borne miR-216b that modulates levels of miR-216b in zygotes and K-RAS in two-cell embryos. This modulation might regulate early development by interfering with the first cleavage and blastocyst quality.
Conflict of interest statement
M.F.S.F. is employed by Alta Genetics; M.B.R.A., R.P.A., T.H.C.D.B., S.A.F.R., C.B., F.V.M., J.C.S., F.P. and E.C.C.C. have nothing to declare.
Figures


References
-
- Vincent P, et al. Bovine semen quality control in artificial insemination centers. Anim. Reprod. 2012;9:153–165.
-
- Oliveira LZ, et al. Assessment of field fertility and several in vitro sperm characteristics following the use of different Angus sires in a timed-AI program with suckled Nelore cows. Livest. Sci. 2012;146:38–46. doi: 10.1016/j.livsci.2012.02.018. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

