Biochar Amendment Stimulates Utilization of Plant-Derived Carbon by Soil Bacteria in an Intercropping System
- PMID: 31316475
- PMCID: PMC6611431
- DOI: 10.3389/fmicb.2019.01361
Biochar Amendment Stimulates Utilization of Plant-Derived Carbon by Soil Bacteria in an Intercropping System
Abstract
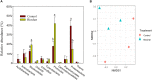
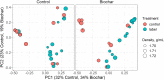
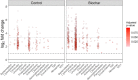
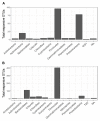
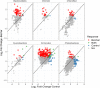
Plant-derived carbon (C) is considered fundamental to understand the interaction between rhizosphere microbes and plants in terrestrial ecosystems. Biochar soil amendment may enhance plant performance via changing soil properties or microbial diversity in the rhizosphere. However, our knowledge of how plant-microbiome associations respond to biochar amendment remains rather limited. Herein, 13CO2 steady-state labeling combined with DNA stable-isotope probing was used to characterize soil bacterial communities in the rhizosphere contributing to the utilization of plant-derived C. The diversity of bacteria active in the utilization of root exudates was determined after biochar amendment in a legume-based intercropping system (Vicia faba L., with Zea mays L.). The results showed the biochar application not only changed the bacterial community structure and diversity in the rhizosphere, but also altered bacterial members actively assimilating plant-derived C. There were more labeled species in the biochar-amended soils than the control soils. Compared with the control, the biochar amendment increased the relative abundances of Firmicutes and Bacteroidetes members (i.e., Bacillus, Clostridium, Sporomusa, Desulfosporosinus, and Alicyclobacillus) while decreasing the abundances of Proteobacteria members (e.g., Methylobacterium and Sphingomonas) utilizing plant-derived C. In contrast, slow-growing species of the phyla Acidobacteria, Planctomycetes, and Gemmatimonadetes were barely labeled. The bacteria found stimulated by the biochar amendment are known for their ability to fix nitrogen, solubilize phosphorus, or reduce iron and sulfur, which may potentially contribute to the "biochar effect" in the rhizosphere. This study is the first to provide empirical evidence that biochar amendment can alter the soil bacterial community assimilating plant-derived C; this may have consequences for nutrient cycling and improving plant performance in intercropping systems.
Keywords: 13CO2 steady-state labeling; biochar; intercropping; plant-derived carbon; rhizosphere microbes; stable-isotope probing.
Figures


Similar articles
-
Influence of reductive soil disinfestation or biochar amendment on bacterial communities and their utilization of plant-derived carbon in the rhizosphere of tomato.Appl Microbiol Biotechnol. 2021 Jan;105(2):815-825. doi: 10.1007/s00253-020-11036-6. Epub 2021 Jan 2. Appl Microbiol Biotechnol. 2021. PMID: 33386895
-
[Effects of Selected Biochars Application on the Microbial Community Structures and Diversities in the Rhizosphere of Water Spinach (Ipomoea aquatica Forssk.) Irrigated with Reclaimed Water].Huan Jing Ke Xue. 2020 Dec 8;41(12):5636-5647. doi: 10.13227/j.hjkx.202006087. Huan Jing Ke Xue. 2020. PMID: 33374081 Chinese.
-
Effect of straw biochar amendment on tobacco growth, soil properties, and rhizosphere bacterial communities.Sci Rep. 2021 Oct 20;11(1):20727. doi: 10.1038/s41598-021-00168-y. Sci Rep. 2021. PMID: 34671040 Free PMC article.
-
Biochar amendment improves crop production in problem soils: A review.J Environ Manage. 2019 Feb 15;232:8-21. doi: 10.1016/j.jenvman.2018.10.117. Epub 2018 Nov 20. J Environ Manage. 2019. PMID: 30466010 Review.
-
The Role of Biochar in Reducing the Bioavailability and Migration of Persistent Organic Pollutants in Soil-Plant Systems: A Review.Bull Environ Contam Toxicol. 2020 Feb;104(2):157-165. doi: 10.1007/s00128-019-02779-8. Epub 2020 Jan 2. Bull Environ Contam Toxicol. 2020. PMID: 31898750 Review.
Cited by
-
Free-living bacteria stimulate sugarcane growth traits and edaphic factors along soil depth gradients under contrasting fertilization.Sci Rep. 2023 Apr 18;13(1):6288. doi: 10.1038/s41598-022-25807-w. Sci Rep. 2023. PMID: 37072423 Free PMC article.
-
Genotype Combinations Drive Variability in the Microbiome Configuration of the Rhizosphere of Maize/Bean Intercropping System.Int J Mol Sci. 2024 Jan 20;25(2):1288. doi: 10.3390/ijms25021288. Int J Mol Sci. 2024. PMID: 38279288 Free PMC article.
-
Influence of reductive soil disinfestation or biochar amendment on bacterial communities and their utilization of plant-derived carbon in the rhizosphere of tomato.Appl Microbiol Biotechnol. 2021 Jan;105(2):815-825. doi: 10.1007/s00253-020-11036-6. Epub 2021 Jan 2. Appl Microbiol Biotechnol. 2021. PMID: 33386895
-
Pyrolysis Temperature vs. Application Rate of Biochar Amendments: Impacts on Soil Microbiota and Metribuzin Degradation.Int J Mol Sci. 2023 Jul 6;24(13):11154. doi: 10.3390/ijms241311154. Int J Mol Sci. 2023. PMID: 37446332 Free PMC article.
-
Distinct biophysical and chemical mechanisms governing sucrose mineralization and soil organic carbon priming in biochar amended soils: evidence from 10 years of field studies.Biochar. 2024;6(1):52. doi: 10.1007/s42773-024-00327-0. Epub 2024 May 22. Biochar. 2024. PMID: 38799721 Free PMC article.
References
-
- Abel S., Peters A., Trinks S., Schonsky H., Facklam M., Wessolek G. (2013). Impact of biochar and hydrochar addition on water retention and water repellency of sandy soil. Geoderma 202 183–191. 10.1016/j.geoderma.2013.03.003 - DOI
-
- Badri D. V., Chaparro J. M., Zhang R., Shen Q., Vivanco J. M. (2013). Application of natural blends of phytochemicals derived from the root exudates of Arabidopsis to the soil reveal that phenolic-related compounds predominantly modulate the soil microbiome. J. Biol. Chem. 288 4502–4512. 10.1074/jbc.M112.433300 - DOI - PMC - PubMed
-
- Bao S. D. (1999). Analytical Methods of Soil Agrochemistry (in Chinese). Beijing: China Agricultural Press.
LinkOut - more resources
Full Text Sources

