Key Features Relevant to Select Antigens and TCR From the MHC-Mismatched Repertoire to Treat Cancer
- PMID: 31316521
- PMCID: PMC6611213
- DOI: 10.3389/fimmu.2019.01485
Key Features Relevant to Select Antigens and TCR From the MHC-Mismatched Repertoire to Treat Cancer
Abstract
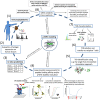
Adoptive transfer of T cells transgenic for tumor-reactive T-cell receptors (TCR) is an attractive immunotherapeutic approach. However, clinical translation is so far limited due to challenges in the identification of suitable target antigens as well as TCRs that are concurrent safe and efficient. Definition of key characteristics relevant for effective and specific tumor rejection is essential to improve current TCR-based adoptive T-cell immunotherapies. We here characterized in-depth two TCRs derived from the human leukocyte antigen (HLA)-mismatched allogeneic repertoire targeting two different myeloperoxidase (MPO)-derived peptides presented by the same HLA-restriction element side by side comprising state of the art biochemical and cellular in vitro, in vivo, and in silico experiments. In vitro experiments reveal comparable functional avidities, off-rates, and cytotoxic activities for both TCRs. However, we observed differences especially with respect to cytokine secretion and cross-reactivity as well as in vivo activity. Biochemical and in silico analyses demonstrate different binding qualities of MPO-peptides to the HLA-complex determining TCR qualities. We conclude from our biochemical and in silico analyses of peptide-HLA-binding that rigid and high-affinity binding of peptides is one of the most important factors for isolation of TCRs with high specificity and tumor rejection capacity from the MHC-mismatched repertoire. Based on our results, we developed a workflow for selection of such TCRs with high potency and safety profile suitable for clinical translation.
Keywords: T-cell receptor (TCR); TCR cell therapy; TCR characterization; TCR identification; adoptive T-cell transfer therapy; peptide-MHC modeling (p-MHC modeling); target antigen characterization; trimolecular complex (TCR-p-MHC).
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

