Whole Genome Sequencing: Bridging One-Health Surveillance of Foodborne Diseases
- PMID: 31316960
- PMCID: PMC6610495
- DOI: 10.3389/fpubh.2019.00172
Whole Genome Sequencing: Bridging One-Health Surveillance of Foodborne Diseases
Erratum in
-
Corrigendum: Whole Genome Sequencing: Bridging One-Health Surveillance of Foodborne Diseases.Front Public Health. 2019 Dec 6;7:365. doi: 10.3389/fpubh.2019.00365. eCollection 2019. Front Public Health. 2019. PMID: 31867301 Free PMC article.
Abstract
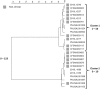
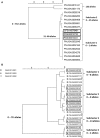
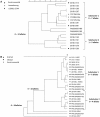
Infections caused by pathogens commonly acquired from consumption of food are not always transmitted by that route. They may also be transmitted through contact to animals, other humans or the environment. Additionally, many outbreaks are associated with food contaminated from these non-food sources. For this reason, such presumed foodborne outbreaks are best investigated through a One Health approach working across human, animal and environmental sectors and disciplines. Outbreak strains or clones that have propagated and continue to evolve in non-human sources and environments often show more sequence variation than observed in typical monoclonal point-source outbreaks. This represents a challenge when using whole genome sequencing (WGS), the new gold standard for molecular surveillance of foodborne pathogens, for outbreak detection and investigation. In this review, using recent examples from outbreaks investigated in the United States (US) some aspects of One Health approaches that have been used successfully to solve such outbreaks are presented. These include using different combinations of flexible WGS based case definition, efficient epidemiological follow-up, traceback, surveillance, and testing of potential food and environmental sources and animal hosts.
Keywords: animals; environment; food; investigation; one health; outbreak; whole genome sequencing (WGS); zoonotic.
Figures


References
-
- Schurch AC, Arredondo-Alonso S, Willems RJL, Goering RV. Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene-based approaches. Clin Microbiol Infect. (2018) 24:350–4. 10.1016/j.cmi.2017.12.016 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources

