Mechanism of Single-Stranded DNA Activation of Recombinase Intein Splicing
- PMID: 31318538
- PMCID: PMC8409432
- DOI: 10.1021/acs.biochem.9b00506
Mechanism of Single-Stranded DNA Activation of Recombinase Intein Splicing
Abstract
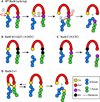
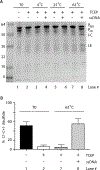
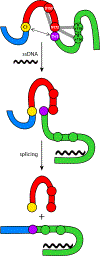
Inteins, or intervening proteins, are mobile genetic elements translated within host polypeptides and removed through protein splicing. This self-catalyzed process breaks two peptide bonds and rejoins the flanking sequences, called N- and C-exteins, with the intein scarlessly escaping the host protein. As these elements have traditionally been viewed as purely selfish genetic elements, recent work has demonstrated that the conditional protein splicing (CPS) of several naturally occurring inteins can be regulated by a variety of environmental cues relevant to the survival of the host organism or crucial to the invading protein function. The RadA recombinase from the archaeon Pyrococcus horikoshii represents an intriguing example of CPS, whereby protein splicing is inhibited by interactions between the intein and host protein C-extein. Single-stranded DNA (ssDNA), a natural substrate of RadA as well as signal that recombinase activity is needed by the cell, dramatically improves the splicing rate and accuracy. Here, we investigate the mechanism by which ssDNA exhibits this influence and find that ssDNA strongly promotes a specific step of the splicing reaction, cyclization of the terminal asparagine of the intein. Interestingly, inhibitory interactions between the host protein and intein that block splicing localize to this asparagine, suggesting that ssDNA binding alleviates this inhibition to promote splicing. We also find that ssDNA directly influences the position of catalytic nucleophiles required for protein splicing, implying that ssDNA promotes assembly of the intein active site. This work advances our understanding of how ssDNA accelerates RadA splicing, providing important insights into this intriguing example of CPS.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Paulus H (2001) Inteins as enzymes. Bioorg. Chem. 29, 119–129. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

