Car-Parrinello Monitor for More Robust Born-Oppenheimer Molecular Dynamics
- PMID: 31318557
- PMCID: PMC9749491
- DOI: 10.1021/acs.jctc.9b00439
Car-Parrinello Monitor for More Robust Born-Oppenheimer Molecular Dynamics
Abstract
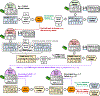
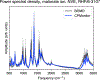
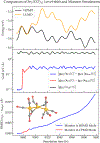
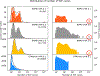
Born-Oppenheimer molecular dynamics (BOMD) is a promising simulation method for exploring the possible reaction pathways of a chemical system, but one significant challenge is the increased difficulty of converging the self-consistent field (SCF) calculation that often accompanies the breaking and forming of chemical bonds. To address this challenge, we developed an enhancement to the BOMD simulation method called the Car-Parrinello monitor (CPMonitor) that uses Car-Parrinello molecular dynamics (CPMD) to recover from SCF convergence failures. CPMonitor works by detecting SCF convergence failures in BOMD and switching to a CPMD Hamiltonian to propagate through the region of configuration space where the SCF calculation is unable to converge, then switching back to BOMD when good convergence behavior is re-established. We present a series of simulation studies that use CPMonitor, including detailed studies of the thermodynamic and dynamical properties of simple systems, as well as ab initio nanoreactor simulations containing transition metal atoms that were previously not possible to simulate using standard BOMD methods. Our studies show that CPMonitor can make BOMD simulations robust to SCF convergence difficulties and improve simulation performance and stability in reaction discovery applications.
Figures



References
-
- Payne MC; Teter MP; Allan DC; Arias TA; Joannopoulos JD Iterative minimization techniques for ab initio total-energy calculations: molecular dynamics and conjugate gradients. Rev. Mod. Phys 1992, 64, 1045–1097.
-
- Thompson DL Modern Methods for Multidimensional Dynamics Computations in Chemistry; WORLD SCIENTIFIC, 1998.
-
- Tuckerman ME Ab initio molecular dynamics: basic concepts, current trends and novel applications. J. Phys.-Condens. Mat 2002, 14, R1297–R1355.
-
- Leforestier C. Classical trajectories using the full ab initio potential energy surface H+CH4→CH4+H. J. Chem. Phys 1978, 68, 4406–4410.
-
- Ishida K; Morokuma K; Komornicki A. The intrinsic reaction coordinate. An ab initio calculation for HNC→HCN and H+CH4→CH4+H. J. Chem. Phys 1977, 66, 2153–2156.
