Effects of Moloney Leukemia Virus 10 Protein on Hepatitis B Virus Infection and Viral Replication
- PMID: 31319455
- PMCID: PMC6669478
- DOI: 10.3390/v11070651
Effects of Moloney Leukemia Virus 10 Protein on Hepatitis B Virus Infection and Viral Replication
Abstract
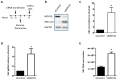
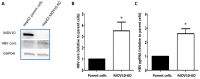
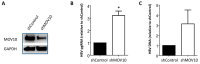
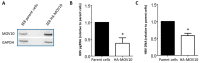
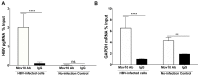
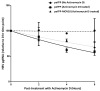
Moloney leukemia virus 10 (MOV10) is an RNA helicase that has been shown to affect the replication of several viruses. The effect of MOV10 on Hepatitis B virus (HBV) infection is not known and its role on the replication of this virus is poorly understood. We investigated the effect of MOV10 down-regulation and MOV10 over-expression on HBV in a variety of cell lines, as well as in an infection system using a replication competent virus. We report that MOV10 down-regulation, using siRNA, shRNA, and CRISPR/Cas9 gene editing technology, resulted in increased levels of HBV DNA, HBV pre-genomic RNA, and HBV core protein. In contrast, MOV10 over-expression reduced HBV DNA, HBV pre-genomic RNA, and HBV core protein. These effects were consistent in all tested cell lines, providing strong evidence for the involvement of MOV10 in the HBV life cycle. We demonstrated that MOV10 does not interact with HBV-core. However, MOV10 binds HBV pgRNA and this interaction does not affect HBV pgRNA decay rate. We conclude that the restriction of HBV by MOV10 is mediated through effects at the level of viral RNA.
Keywords: gene expression; hepatitis B virus; host factors; moloney leukemia virus 10.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

