A Photosynthesis-Specific Rubredoxin-Like Protein Is Required for Efficient Association of the D1 and D2 Proteins during the Initial Steps of Photosystem II Assembly
- PMID: 31320483
- PMCID: PMC6751121
- DOI: 10.1105/tpc.19.00155
A Photosynthesis-Specific Rubredoxin-Like Protein Is Required for Efficient Association of the D1 and D2 Proteins during the Initial Steps of Photosystem II Assembly
Abstract
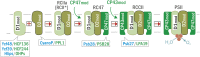
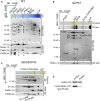
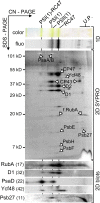
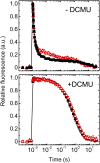
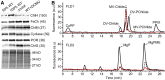
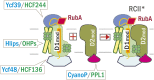
Oxygenic photosynthesis relies on accessory factors to promote the assembly and maintenance of the photosynthetic apparatus in the thylakoid membranes. The highly conserved membrane-bound rubredoxin-like protein RubA has previously been implicated in the accumulation of both PSI and PSII, but its mode of action remains unclear. Here, we show that RubA in the cyanobacterium Synechocystis sp PCC 6803 is required for photoautotrophic growth in fluctuating light and acts early in PSII biogenesis by promoting the formation of the heterodimeric D1/D2 reaction center complex, the site of primary photochemistry. We find that RubA, like the accessory factor Ycf48, is a component of the initial D1 assembly module as well as larger PSII assembly intermediates and that the redox-responsive rubredoxin-like domain is located on the cytoplasmic surface of PSII complexes. Fusion of RubA to Ycf48 still permits normal PSII assembly, suggesting a spatiotemporal proximity of both proteins during their action. RubA is also important for the accumulation of PSI, but this is an indirect effect stemming from the downregulation of light-dependent chlorophyll biosynthesis induced by PSII deficiency. Overall, our data support the involvement of RubA in the redox control of PSII biogenesis.
© 2019 American Society of Plant Biologists. All rights reserved.
Figures






References
-
- Bečková M., Gardian Z., Yu J., Koník P., Nixon P.J., Komenda J. (2017). Association of Psb28 and Psb27 proteins with PSII-PSI supercomplexes upon exposure of Synechocystis sp. PCC 6803 to high light. Mol. Plant 10: 62–72. - PubMed
-
- Boehm M., Yu J., Reisinger V., Bečkova M., Eichacker L.A., Schlodder E., Komenda J., Nixon P.J. (2012). Subunit composition of CP43-less photosystem II complexes of Synechocystis sp. PCC 6803: Implications for the assembly and repair of photosystem II. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 3444–3454. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

