Exosomes regulate neurogenesis and circuit assembly
- PMID: 31320591
- PMCID: PMC6689941
- DOI: 10.1073/pnas.1902513116
Exosomes regulate neurogenesis and circuit assembly
Abstract
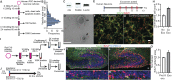
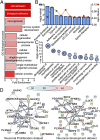
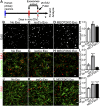
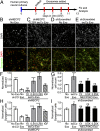
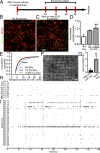
Exosomes are thought to be released by all cells in the body and to be involved in intercellular communication. We tested whether neural exosomes can regulate the development of neural circuits. We show that exosome treatment increases proliferation in developing neural cultures and in vivo in dentate gyrus of P4 mouse brain. We compared the protein cargo and signaling bioactivity of exosomes released by hiPSC-derived neural cultures lacking MECP2, a model of the neurodevelopmental disorder Rett syndrome, with exosomes released by isogenic rescue control neural cultures. Quantitative proteomic analysis indicates that control exosomes contain multiple functional signaling networks known to be important for neuronal circuit development. Treating MECP2-knockdown human primary neural cultures with control exosomes rescues deficits in neuronal proliferation, differentiation, synaptogenesis, and synchronized firing, whereas exosomes from MECP2-deficient hiPSC neural cultures lack this capability. These data indicate that exosomes carry signaling information required to regulate neural circuit development.
Keywords: Rett syndrome; exosomes; extracellular vesicle; neuronal development; synaptogenesis.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
Conflict of interest statement: The authors declare a conflict of interest. A.R.M. is a cofounder and has equity interest in TISMOO, a company dedicated to genetic analysis focusing on therapeutic applications customized for autism spectrum disorder and other neurological disorders with genetic origins. The terms of this arrangement have been reviewed and approved by the University of California San Diego, in accordance with its conflict of interest policies. The remaining authors declare that they have no conflict of interest.
Figures
Comment in
-
Promotion of synaptogenesis and neural circuit development by exosomes.Ann Transl Med. 2019 Dec;7(Suppl 8):S323. doi: 10.21037/atm.2019.09.154. Ann Transl Med. 2019. PMID: 32016041 Free PMC article. No abstract available.
References
-
- Chiasserini D., et al. , Proteomic analysis of cerebrospinal fluid extracellular vesicles: A comprehensive dataset. J. Proteomics 106, 191–204 (2014). - PubMed
-
- Coleman B. M., Hill A. F., Extracellular vesicles–Their role in the packaging and spread of misfolded proteins associated with neurodegenerative diseases. Semin. Cell Dev. Biol. 40, 89–96 (2015). - PubMed

