Novel hepatitis D-like agents in vertebrates and invertebrates
- PMID: 31321078
- PMCID: PMC6628682
- DOI: 10.1093/ve/vez021
Novel hepatitis D-like agents in vertebrates and invertebrates
Abstract
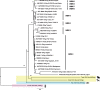
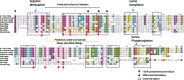
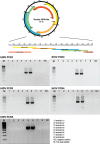
Hepatitis delta virus (HDV) is the smallest known RNA virus, encoding a single protein. Until recently, HDV had only been identified in humans, where it is strongly associated with co-infection with hepatitis B virus (HBV). However, the recent discovery of HDV-like viruses in metagenomic samples from birds and snakes suggests that this virus has a far longer evolutionary history. Herein, using additional meta-transcriptomic data, we show that highly divergent HDV-like viruses are also present in fish, amphibians, and invertebrates, with PCR and Sanger sequencing confirming the presence of the invertebrate HDV-like viruses. Notably, the novel viruses identified here share genomic features characteristic of HDV, such as a circular genome of only approximately 1.7 kb in length, and self-complementary, unbranched rod-like structures. Coiled-coil domains, leucine zippers, conserved residues with essential biological functions, and isoelectronic points similar to those in the human hepatitis delta virus antigens (HDAgs) were also identified in the putative non-human viruses. Importantly, none of these novel HDV-like viruses were associated with hepadnavirus infection, supporting the idea that the HDV-HBV association may be specific to humans. Collectively, these data not only broaden our understanding of the diversity and host range of HDV, but also shed light on its origin and evolutionary history.
Keywords: evolution; fish; hepatitis D virus; meta-transcriptomics; phylogeny; termites.
Figures


References
-
- Anisimova M., Yang Z. (2004) ‘Molecular Evolution of the Hepatitis Delta Virus Antigen Gene: Recombination or Positive Selection?’, Journal of Molecular Evolution, 59: 815–26. - PubMed
-
- Brazas R., Ganem D. (1996) ‘A Cellular Homolog of Hepatitis Delta Antigen: Implications for Viral Replication and Evolution’, Science, 274: 90–4. - PubMed
-
- Buchfink B., Xie C., Huson D. H. (2015) ‘Fast and Sensitive Protein Alignment Using DIAMOND’, Nature Methods, 12: 59–60. - PubMed
-
- Bushnell B. (2016) BBMap Short-Read Aligner, and Other Bioinformatics Toolshttps://sourceforge.net/projects/bbmap/ accessed 8 Nov 2018.
LinkOut - more resources
Full Text Sources

