Molecular identification and genetic variation of Alternaria species isolated from tomatoes using ITS1 sequencing and inter simple sequence repeat methods
- PMID: 31321331
- PMCID: PMC6626712
- DOI: 10.18502/cmm.5.2.1154
Molecular identification and genetic variation of Alternaria species isolated from tomatoes using ITS1 sequencing and inter simple sequence repeat methods
Abstract
Background and purpose: Alternaria is one of the most abundant fungi that exists in numerous places around the world. This saprophytic fungus causes diseases in plants and accounts for the spoilage of cereals in warehouses. The aim of this study was to identify Alternaria isolates based on their morphological characteristics and internal transcribed spacer ribosomal RNA (ITS rRNA) sequencing method. To this end, genetic diversity in the isolates was also examined using inter simple sequence repeat (ISSR) markers.
Materials and methods: To conduct this research, a total of 60 tomato samples with black spots were collected from supermarkets in Khorramabad City, Iran, in the winter of 2017. The specimens were cultured on a potato dextrose agar medium. After the purification of the fungus by the single-spore method, the identification of the species was carried out using morphological characteristics and ITS rRNA sequencing by polymerase chain reaction. The genetic diversity of the identified species with four primers was evaluated using the ISSR marker.
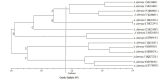
Results: Based on the sequencing of the ITS1 region, all the isolates were identified as A. alternata. Cluster diagrams for the ISSR marker were classified into six distinct groups. The mean polymorphism information content was obtained as 0.35, indicating the effectiveness of the primers in the separation of the isolates.
Conclusion: The sequencing of ITS1 led to the identification of Alternaria species that are morphologically similar. The production of various mycotoxins by A . Alternata species leads to the contamination of livestock and human food. Regarding this, the investigation of the genetic diversity of A. alternata species using the ISSR marker would facilitate the identification of suitable and effective strategies for controlling the fungal and mycotoxin contamination of human nutrition.
Keywords: Alternaria alternata; ITS rRNA; Inter simple sequence repeat methods; Tomato.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures


References
-
- MacDonald S, Prickett T, Wildey K, Chan D. Survey of ochratoxin A and deoxynivalenol in stored grains from the 1999 harvest in the UK. Food Addit Contam. 2004;21(2)::172–81. - PubMed
-
- Pitt JI, Hocking AD. Fungi and food spoilage. New South Wales, Australia: Blackie Academic & Professional; 1997.
-
- Tutelyan VA. Deoxynivalenol in cereals in Russia. Toxicol Lett. 2004;153(1):173–9. - PubMed
-
- Thomma BP. Alternaria spp from general saprophyte to specific parasite. Mol Plant Pathol. 2003;4(4)::225–36. - PubMed
-
- Samson RA, Houbraken J, Thrane U, Frisvad JC, Andersen B. Food and indoor fungi. Netherlands. CBS-KNAW Fungal Biodiversity Centre: 2010.
LinkOut - more resources
Full Text Sources
Other Literature Sources
