Repeat-associated RNA structure and aberrant splicing
- PMID: 31323433
- PMCID: PMC7099610
- DOI: 10.1016/j.bbagrm.2019.07.006
Repeat-associated RNA structure and aberrant splicing
Abstract
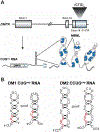
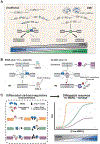
Over 30 hereditary disorders attributed to the expansion of microsatellite repeats have been identified. Despite variant nucleotide content, number of consecutive repeats, and different locations in the genome, many of these diseases have pathogenic RNA gain-of-function mechanisms. The repeat-containing RNAs can form structures in vitro predicted to contribute to the disease through assembly of intracellular RNA aggregates termed foci. The expanded repeat RNAs within these foci sequester RNA binding proteins (RBPs) with important roles in the regulation of RNA metabolism, most notably alternative splicing (AS). These deleterious interactions lead to downstream alterations in transcriptome-wide AS directly linked with disease symptoms. This review summarizes existing knowledge about the association between the repeat RNA structures and RBPs as well as the resulting aberrant AS patterns, specifically in the context of myotonic dystrophy. The connection between toxic, structured RNAs and dysregulation of AS in other repeat expansion diseases is also discussed. This article is part of a Special Issue entitled: RNA structure and splicing regulation edited by Francisco Baralle, Ravindra Singh and Stefan Stamm.
Keywords: Alternative splicing; Muscleblind-like (MBNL); Myotonic dystrophy; RNA binding proteins; RNA structure; Repeat expansion diseases.
Copyright © 2019 Elsevier B.V. All rights reserved.
Figures
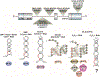
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

