Chromatin folding and nuclear architecture: PRC1 function in 3D
- PMID: 31323466
- PMCID: PMC6859790
- DOI: 10.1016/j.gde.2019.06.006
Chromatin folding and nuclear architecture: PRC1 function in 3D
Abstract
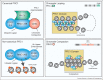
Embryonic development requires the intricate balance between the expansion and specialisation of defined cell types in time and space. The gene expression programmes that underpin this balance are regulated, in part, by modulating the chemical and structural state of chromatin. Polycomb repressive complexes (PRCs), a family of essential developmental regulators, operate at this level to stabilise or perpetuate a repressed but transcriptionally poised chromatin configuration. This dynamic state is required to control the timely initiation of productive gene transcription during embryonic development. The two major PRCs cooperate to target the genome, but it is PRC1 that appears to be the primary effector that controls gene expression. In this review I will discuss recent findings relating to how PRC1 alters chromatin accessibility, folding and global 3D nuclear organisation to control gene transcription.
Copyright © 2019 The Author. Published by Elsevier Ltd.. All rights reserved.
Figures

References
-
- Gilbert N., Boyle S., Fiegler H., Woodfine K., Carter N.P., Bickmore W.A. Chromatin architecture of the human genome gene-rich domains are enriched in open chromatin fibers. Cell. 2004;118:555–566. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

