Computational Processing and Quality Control of Hi-C, Capture Hi-C and Capture-C Data
- PMID: 31323892
- PMCID: PMC6678864
- DOI: 10.3390/genes10070548
Computational Processing and Quality Control of Hi-C, Capture Hi-C and Capture-C Data
Abstract
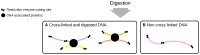
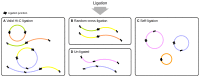
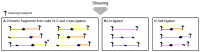
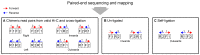
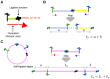
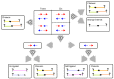
Hi-C, capture Hi-C (CHC) and Capture-C have contributed greatly to our present understanding of the three-dimensional organization of genomes in the context of transcriptional regulation by characterizing the roles of topological associated domains, enhancer promoter loops and other three-dimensional genomic interactions. The analysis is based on counts of chimeric read pairs that map to interacting regions of the genome. However, the processing and quality control presents a number of unique challenges. We review here the experimental and computational foundations and explain how the characteristics of restriction digests, sonication fragments and read pairs can be exploited to distinguish technical artefacts from valid read pairs originating from true chromatin interactions.
Keywords: Hi-C; capture Hi-C; processing; quality control.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Lieberman-Aiden E., Van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O., et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

