Gene expression profile of human T cells following a single stimulation of peripheral blood mononuclear cells with anti-CD3 antibodies
- PMID: 31324145
- PMCID: PMC6642599
- DOI: 10.1186/s12864-019-5967-8
Gene expression profile of human T cells following a single stimulation of peripheral blood mononuclear cells with anti-CD3 antibodies
Abstract
Background: Anti-CD3 immunotherapy was initially approved for clinical use for renal transplantation rejection prevention. Subsequently, new generations of anti-CD3 antibodies have entered clinical trials for a broader spectrum of therapeutic applications, including cancer and autoimmune diseases. Despite their extensive use, little is known about the exact mechanism of these molecules, except that they are able to activate T cells, inducing an overall immunoregulatory and tolerogenic behavior. To better understand the effects of anti-CD3 antibodies on human T cells, PBMCs were stimulated, and then, we performed RNA-seq assays of enriched T cells to assess changes in their gene expression profiles. In this study, three different anti-CD3 antibodies were used for the stimulation: two recombinant antibody fragments, namely, a humanized and a chimeric FvFc molecule, and the prototype mouse mAb OKT3.
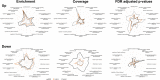
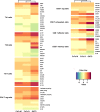
Results: Gene Ontology categories and individual immunoregulatory markers were compared, suggesting a similarity in modulated gene sets, mainly those for immunoregulatory and inflammatory terms. Upregulation of interleukin receptors, such as IL2RA, IL1R, IL12RB2, IL18R1, IL21R and IL23R, and of inhibitory molecules, such as FOXP3, CTLA4, TNFRSF18, LAG3 and PDCD1, were also observed, suggesting an inhibitory and exhausted phenotype.
Conclusions: We used a deep transcriptome sequencing method for comparing three anti-CD3 antibodies in terms of Gene Ontology enrichment and immunological marker expression. The present data showed that both recombinant antibodies induced a compatible expression profile, suggesting that they might be candidates for a closer evaluation with respect to their therapeutic value. Moreover, the proposed methodology is amenable to be more generally applied for molecular comparison of cell receptor dependent antibody therapy.
Keywords: Anti-CD3; Antibody engineering; Antibody therapy; RNA-seq; Regulatory T cells.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- Cosimi AB, Urton B, Colvin RB, Goldstein G, Laquaglia MP, Tolkoff-Rubin N, et al. Treatment of acute renal allograft rejection with OKT3 monoclonal antibody. Transplantation. 1981;32(6):535–539. - PubMed
-
- Chatenoud L, Primo J, Bach JF. CD3 antibody-induced dominant self tolerance in overtly diabetic NOD mice. J Immunol. 1997;158(6):2947–2954. - PubMed
-
- Chatenoud L, Bluestone JA. CD3-specific antibodies: a portal to the treatment of autoimmunity. Nat Rev Immunol. 2007;7(8):622–632. - PubMed
-
- Utset TO, Auger JA, Peace D, Zivin RA, Xu D, Jolliffe L, et al. Modified anti-CD3 therapy in psoriatic arthritis: a phase I/II clinical trial. J Rheumatol. 2002;29(9):1907–1913. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

