Amino Acids and Ribose: Drivers of Protein and RNA Fermentation by Ingested Bacteria of a Primitive Gut Ecosystem
- PMID: 31324631
- PMCID: PMC6752017
- DOI: 10.1128/AEM.01297-19
Amino Acids and Ribose: Drivers of Protein and RNA Fermentation by Ingested Bacteria of a Primitive Gut Ecosystem
Abstract
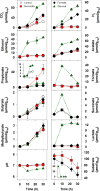
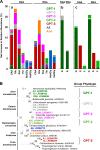
Earthworms are among the most primitive animals and are of fundamental importance to the turnover of organic matter in the terrestrial biosphere. These invertebrates ingest materials that are colonized by microbes, some of which are subject to disruption by the crop/gizzard or other lytic events during gut passage. Protein and RNA are dominant polymers of disrupted microbial cells, and these biopolymers facilitate robust fermentations by surviving ingested bacteria. To further resolve these fermentations, amino acids and ribose (as fermentable constituents of protein and RNA, respectively) were evaluated as potential drivers of fermentation in gut content of the model earthworm Lumbricus terrestris (taxa were examined with 16S rRNA-based analyses). Of eight amino acids tested, glutamate, aspartate, and threonine were most stimulatory and yielded dissimilar fermentations facilitated by contrasting taxa (e.g., glutamate stimulated the Fusobacteriaceae and yielded H2 and formate, whereas aspartate stimulated the Aeromonadaceae and yielded succinate and propionate). A marginal Stickland fermentation was associated with the Peptostreptococcaceae and Lachnospiraceae Ribose fermentation yielded a complex product profile facilitated primarily by the Aeromonadaceae The transient nature of succinate was linked to its decarboxylation to propionate and the Fusobacteriaceae, whereas the transient nature of formate was linked to formate-hydrogen lyase activity and the Peptostreptococcaceae These findings reinforce the likelihood that (i) the animal host and hosted fermentative bacteria compete for the constituents of protein and RNA in the alimentary canal and (ii) diverse gut fermenters engaged in the fermentation of these constituents produce products that can be utilized by earthworms.IMPORTANCE Animal health is linked to gut ecosystems whose primary function is normally the digestion of dietary matter. Earthworms are representative of one of the oldest known animal lineages and, despite their primitive nature, have unique environmental impact by virtue of their dietary consumption of their habitat, i.e., soil-associated matter. A resident gut community is a hallmark of many gut ecosystems of evolutionarily more advanced animals, but the alimentary canal of earthworms is dominated by ingested transient soil microbes. Protein and RNA are (i) the primary organic components of microbial cells that are subject to lysis during gut passage and (ii) fermentable dietary substrates in the alimentary canal. This study examined the gut-associated fermentation of constituents of these biopolymers to determine how their fermentation is integrated to the microbiological dynamics of the gut and might contribute to earthworm-linked transformations of organic matter in the terrestrial biosphere.
Keywords: amino acid fermentation; anaerobes; gut ecosystem; invertebrate microbiology.
Copyright © 2019 Zeibich et al.
Figures





References
-
- Morris SC, Peel JS. 2008. The earliest annelids: lower Cambrian polychaetes from the Sirius Passet Lagerstätte, Peary Land, North Greenland. Acta Palaeontol Pol 53:137–148. doi:10.4202/app.2008.0110. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

