Plastics: Environmental and Biotechnological Perspectives on Microbial Degradation
- PMID: 31324632
- PMCID: PMC6752018
- DOI: 10.1128/AEM.01095-19
Plastics: Environmental and Biotechnological Perspectives on Microbial Degradation
Abstract
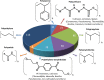
Plastics are widely used in the global economy, and each year, at least 350 to 400 million tons are being produced. Due to poor recycling and low circular use, millions of tons accumulate annually in terrestrial or marine environments. Today it has become clear that plastic causes adverse effects in all ecosystems and that microplastics are of particular concern to our health. Therefore, recent microbial research has addressed the question of if and to what extent microorganisms can degrade plastics in the environment. This review summarizes current knowledge on microbial plastic degradation. Enzymes available act mainly on the high-molecular-weight polymers of polyethylene terephthalate (PET) and ester-based polyurethane (PUR). Unfortunately, the best PUR- and PET-active enzymes and microorganisms known still have moderate turnover rates. While many reports describing microbial communities degrading chemical additives have been published, no enzymes acting on the high-molecular-weight polymers polystyrene, polyamide, polyvinylchloride, polypropylene, ether-based polyurethane, and polyethylene are known. Together, these polymers comprise more than 80% of annual plastic production. Thus, further research is needed to significantly increase the diversity of enzymes and microorganisms acting on these polymers. This can be achieved by tapping into the global metagenomes of noncultivated microorganisms and dark matter proteins. Only then can novel biocatalysts and organisms be delivered that allow rapid degradation, recycling, or value-added use of the vast majority of most human-made polymers.
Keywords: PET; cutinase; microbial plastic degradation; polyamides; polyethylene; polyethylene terephthalate; polypropylene; polystyrene; polyurethane; polyvinylchloride.
Copyright © 2019 Danso et al.
Figures

References
-
- PlasticsEurope. 2018. PlasticsEurope, plastics—the facts 2018: an analysis of European plastics production, demand and waste data. PlasticsEurope, Brussels, Belgium.
-
- Ellen MacArthur Foundation. 2017. The new plastics economy: rethinking the future of plastics and catalysing action. Ellen MacArthur Foundation, Cowes, United Kingdom.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

