RNA structure drives interaction with proteins
- PMID: 31324771
- PMCID: PMC6642211
- DOI: 10.1038/s41467-019-10923-5
RNA structure drives interaction with proteins
Abstract
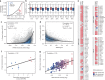
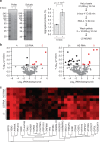
The combination of high-throughput sequencing and in vivo crosslinking approaches leads to the progressive uncovering of the complex interdependence between cellular transcriptome and proteome. Yet, the molecular determinants governing interactions in protein-RNA networks are not well understood. Here we investigated the relationship between the structure of an RNA and its ability to interact with proteins. Analysing in silico, in vitro and in vivo experiments, we find that the amount of double-stranded regions in an RNA correlates with the number of protein contacts. This relationship -which we call structure-driven protein interactivity- allows classification of RNA types, plays a role in gene regulation and could have implications for the formation of phase-separated ribonucleoprotein assemblies. We validate our hypothesis by showing that a highly structured RNA can rearrange the composition of a protein aggregate. We report that the tendency of proteins to phase-separate is reduced by interactions with specific RNAs.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

