Deletion of a Csf1r enhancer selectively impacts CSF1R expression and development of tissue macrophage populations
- PMID: 31324781
- PMCID: PMC6642117
- DOI: 10.1038/s41467-019-11053-8
Deletion of a Csf1r enhancer selectively impacts CSF1R expression and development of tissue macrophage populations
Abstract
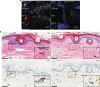
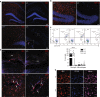
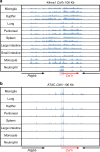
The proliferation, differentiation and survival of mononuclear phagocytes depend on signals from the receptor for macrophage colony-stimulating factor, CSF1R. The mammalian Csf1r locus contains a highly conserved super-enhancer, the fms-intronic regulatory element (FIRE). Here we show that genomic deletion of FIRE in mice selectively impacts CSF1R expression and tissue macrophage development in specific tissues. Deletion of FIRE ablates macrophage development from murine embryonic stem cells. Csf1rΔFIRE/ΔFIRE mice lack macrophages in the embryo, brain microglia and resident macrophages in the skin, kidney, heart and peritoneum. The homeostasis of other macrophage populations and monocytes is unaffected, but monocytes and their progenitors in bone marrow lack surface CSF1R. Finally, Csf1rΔFIRE/ΔFIRE mice are healthy and fertile without the growth, neurological or developmental abnormalities reported in Csf1r-/- rodents. Csf1rΔFIRE/ΔFIRE mice thus provide a model to explore the homeostatic, physiological and immunological functions of tissue-specific macrophage populations in adult animals.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

