PI3K/mTOR inhibition of IDH1 mutant glioma leads to reduced 2HG production that is associated with increased survival
- PMID: 31324855
- PMCID: PMC6642106
- DOI: 10.1038/s41598-019-47021-x
PI3K/mTOR inhibition of IDH1 mutant glioma leads to reduced 2HG production that is associated with increased survival
Abstract
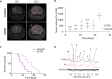
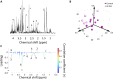
70-90% of low-grade gliomas and secondary glioblastomas are characterized by mutations in isocitrate dehydrogenase 1 (IDHmut). IDHmut produces the oncometabolite 2-hydroxyglutarate (2HG), which drives tumorigenesis in these tumors. The phosphoinositide-3-kinase (PI3K)/mammalian target of rapamycin (mTOR) pathway represents an attractive therapeutic target for IDHmut gliomas, but noninvasive indicators of drug target modulation are lacking. The goal of this study was therefore to identify magnetic resonance spectroscopy (MRS)-detectable metabolic biomarkers associated with IDHmut glioma response to the dual PI3K/(mTOR) inhibitor XL765. 1H-MRS of two cell lines genetically modified to express IDHmut showed that XL765 induced a significant reduction in several intracellular metabolites including 2HG. Importantly, examination of an orthotopic IDHmut tumor model showed that enhanced animal survival following XL765 treatment was associated with a significant in vivo 1H-MRS detectable reduction in 2HG but not with significant inhibition in tumor growth. Further validation is required, but our results indicate that 2HG could serve as a potential noninvasive MRS-detectable metabolic biomarker of IDHmut glioma response to PI3K/mTOR inhibition.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Thakkar JP, et al. Epidemiologic and molecular prognostic review of glioblastoma. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2014;23:1985–1996. doi: 10.1158/1055-9965.epi-14-0275. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

