Ultra-Rare Genetic Variation in the Epilepsies: A Whole-Exome Sequencing Study of 17,606 Individuals
- PMID: 31327507
- PMCID: PMC6698801
- DOI: 10.1016/j.ajhg.2019.05.020
Ultra-Rare Genetic Variation in the Epilepsies: A Whole-Exome Sequencing Study of 17,606 Individuals
Abstract
Sequencing-based studies have identified novel risk genes associated with severe epilepsies and revealed an excess of rare deleterious variation in less-severe forms of epilepsy. To identify the shared and distinct ultra-rare genetic risk factors for different types of epilepsies, we performed a whole-exome sequencing (WES) analysis of 9,170 epilepsy-affected individuals and 8,436 controls of European ancestry. We focused on three phenotypic groups: severe developmental and epileptic encephalopathies (DEEs), genetic generalized epilepsy (GGE), and non-acquired focal epilepsy (NAFE). We observed that compared to controls, individuals with any type of epilepsy carried an excess of ultra-rare, deleterious variants in constrained genes and in genes previously associated with epilepsy; we saw the strongest enrichment in individuals with DEEs and the least strong in individuals with NAFE. Moreover, we found that inhibitory GABAA receptor genes were enriched for missense variants across all three classes of epilepsy, whereas no enrichment was seen in excitatory receptor genes. The larger gene groups for the GABAergic pathway or cation channels also showed a significant mutational burden in DEEs and GGE. Although no single gene surpassed exome-wide significance among individuals with GGE or NAFE, highly constrained genes and genes encoding ion channels were among the lead associations; such genes included CACNA1G, EEF1A2, and GABRG2 for GGE and LGI1, TRIM3, and GABRG2 for NAFE. Our study, the largest epilepsy WES study to date, confirms a convergence in the genetics of severe and less-severe epilepsies associated with ultra-rare coding variation, and it highlights a ubiquitous role for GABAergic inhibition in epilepsy etiology.
Keywords: burden analysis; epilepsy; epileptic encephalopathy; exome; seizures; sequencing.
Copyright © 2019 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
B.M.N. has roles in the following genomics companies: Deep Genomics (scientific advisory board), Camp4 Therapeutics Corporation (scientific advisory board, consultant), Takeda Pharmaceutical (consultant), Biogen-Consultant Genomics Analytics (advisory panel). D.B.G. has equity interest in Praxis Therapeutics and Q State Biosciences, companies focused on precision medicine in neurodevelopmental diseases. S.F.B. is a consultant to Praxis Therapeutics.
Figures

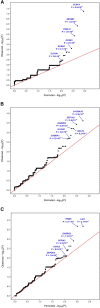
Comment in
-
Epilepsy Genetics: What Once Was Rare, Is Now Common.Epilepsy Curr. 2020 Jun 19;20(4):221-223. doi: 10.1177/1535759720933232. eCollection 2020 Jul-Aug. Epilepsy Curr. 2020. PMID: 34025233 Free PMC article. No abstract available.
References
-
- Fisher R.S., Acevedo C., Arzimanoglou A., Bogacz A., Cross J.H., Elger C.E., Engel J., Jr., Forsgren L., French J.A., Glynn M. ILAE official report: a practical clinical definition of epilepsy. Epilepsia. 2014;55:475–482. - PubMed
-
- Annegers J.F., Hauser W.A., Anderson V.E., Kurland L.T. The risks of seizure disorders among relatives of patients with childhood onset epilepsy. Neurology. 1982;32:174–179. - PubMed
-
- Berkovic S.F., Howell R.A., Hay D.A., Hopper J.L. Epilepsies in twins: genetics of the major epilepsy syndromes. Ann. Neurol. 1998;43:435–445. - PubMed
-
- Helbig I., Scheffer I.E., Mulley J.C., Berkovic S.F. Navigating the channels and beyond: unravelling the genetics of the epilepsies. Lancet Neurol. 2008;7:231–245. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

