Family Clustering of Autoimmune Vitiligo Results Principally from Polygenic Inheritance of Common Risk Alleles
- PMID: 31327509
- PMCID: PMC6698884
- DOI: 10.1016/j.ajhg.2019.06.013
Family Clustering of Autoimmune Vitiligo Results Principally from Polygenic Inheritance of Common Risk Alleles
Abstract
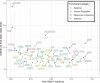
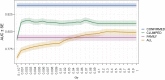
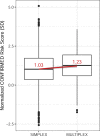
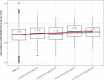
Vitiligo is an autoimmune disease that results in patches of depigmented skin and hair. Previous genome-wide association studies (GWASs) of vitiligo have identified 50 susceptibility loci. Variants at the associated loci are generally common and have individually small effects on risk. Most vitiligo cases are "simplex," where there is no family history of vitiligo, though occasional family clustering of vitiligo occurs, and some "multiplex" families report numerous close affected relatives. Here, we investigate whether simplex and multiplex vitiligo comprise different disease subtypes with different underlying genetic etiologies. We developed and compared the performance of several different vitiligo polygenic risk scores derived from GWAS data. By using the best-performing risk score, we find increased polygenic burden of risk alleles identified by GWAS in multiplex vitiligo cases relative to simplex cases. We additionally find evidence of polygenic transmission of common, low-effect-size risk alleles within multiplex-vitiligo-affected families. Our findings strongly suggest that family clustering of vitiligo involves a high burden of the same common, low-effect-size variants that are relevant in simplex cases. We furthermore find that a variant within the major histocompatibility complex (MHC) class II region contributes disproportionately more to risk in multiplex vitiligo cases than in simplex cases, supporting a special role for adaptive immune triggering in the etiology of multiplex cases. We suggest that genetic risk scores can be a useful tool in analyzing the genetic architecture of clinical disease subtypes and identifying subjects with unusual etiologies for further investigation.
Keywords: autoimmunity; family clustering; genetic architecture; genetic risk score; polygenic inheritance; vitiligo.
Copyright © 2019 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Hafez M., Sharaf L., Abd el-Nabi S.M. The genetics of vitiligo. Acta Derm. Venereol. 1983;63:249–251. - PubMed
-
- Das S., Majumder P.P., Majumdar T., Haldar B., Rao D.J.G.e. Studies on vitiligo. II. Familial aggregation and genetics. Genet. Epidemiol. 1985;2:255–262. - PubMed
-
- Alkhateeb A., Fain P.R., Thody A., Bennett D.C., Spritz R.A. Epidemiology of vitiligo and associated autoimmune diseases in Caucasian probands and their families. Pigment Cell Res. 2003;16:208–214. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

