Divide-and-conquer strategy for large-scale Eulerian solvent excluded surface
- PMID: 31327932
- PMCID: PMC6641554
- DOI: 10.4310/CIS.2018.v18.n4.a5
Divide-and-conquer strategy for large-scale Eulerian solvent excluded surface
Abstract
Motivation: Surface generation and visualization are some of the most important tasks in biomolecular modeling and computation. Eulerian solvent excluded surface (ESES) software provides analytical solvent excluded surface (SES) in the Cartesian grid, which is necessary for simulating many biomolecular electrostatic and ion channel models. However, large biomolecules and/or fine grid resolutions give rise to excessively large memory requirements in ESES construction. We introduce an out-of-core and parallel algorithm to improve the ESES software.
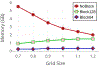
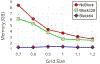
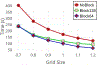
Results: The present approach drastically improves the spatial and temporal efficiency of ESES. The memory footprint and time complexity are analyzed and empirically verified through extensive tests with a large collection of biomolecule examples. Our results show that our algorithm can successfully reduce memory footprint through a straightforward divide-and-conquer strategy to perform the calculation of arbitrarily large proteins on a typical commodity personal computer. On multi-core computers or clusters, our algorithm can reduce the execution time by parallelizing most of the calculation as disjoint subproblems. Various comparisons with the state-of-the-art Cartesian grid based SES calculation were done to validate the present method and show the improved efficiency. This approach makes ESES a robust software for the construction of analytical solvent excluded surfaces.
Availability and implementation: http://weilab.math.msu.edu/ESES.
Figures






References
-
- Asenjo Ana B, Chatterjee Chandrima, Tan Dongyan, Vania DePaoli, Rice William J, Diaz-Avalos Ruben, Silvestry Mariena, and Sosa Hernando, Structural model for tubulin recognition and deformation by kinesin-13 microtubule depolymerases, Cell Reports 3 (2013), no. 3, 759–768. - PubMed
-
- Bates PW, Chen Z, Sun YH, Wei GW, and Zhao S, Geometric and potential driving formation and evolution of biomolecular surfaces, J. Math. Biol 59 (2009), 193–231. - PubMed
-
- Bates PW, Wei Guo-Wei, and Zhao Shan, Minimal molecular surfaces and their applications, Journal of Computational Chemistry 29 (2008), no. 3, 380–391. - PubMed
-
- Blinn J, A generalization of algebraic surface drawing, ACM Transactions on Graphics 1 (1982), no. 3, 235–256.
-
- Chen Minxin and Lu Benzhuo, Tmsmesh: A robust method for molecular surface mesh generation using a trace technique, J Chem. Theory and Comput 7 (2011), 203–212. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
