Genomics of vancomycin-resistant Enterococcus faecium
- PMID: 31329096
- PMCID: PMC6700659
- DOI: 10.1099/mgen.0.000283
Genomics of vancomycin-resistant Enterococcus faecium
Abstract
Vancomycin-resistant Enterococcus faecium (VREfm) is a globally significant public health threat and was listed on the World Health Organization's 2017 list of high-priority pathogens for which new treatments are urgently needed. Treatment options for invasive VREfm infections are very limited, and outcomes are often poor. Whole-genome sequencing is providing important new insights into VREfm evolution, drug resistance and hospital adaptation, and is increasingly being used to track VREfm transmission within hospitals to detect outbreaks and inform infection control practices. This mini-review provides an overview of recent data on the use of genomics to understand and respond to the global problem of VREfm.
Keywords: Enterococcus faecium; antibiotic resistance; whole-genome sequencing.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures
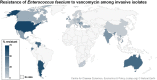
References
-
- Werner G, Coque TM, Hammerum AM, Hope R, Hryniewicz W, et al. Emergence and spread of vancomycin resistance among enterococci in Europe. Euro Surveill Bull Eur Sur Mal Transm Eur Commun Dis Bull. 13 - PubMed
-
- Deshpande LM, Fritsche TR, Moet GJ, Biedenbach DJ, Jones RN. Antimicrobial resistance and molecular epidemiology of vancomycin-resistant enterococci from North America and Europe: a report from the SENTRY antimicrobial surveillance program. Diagn Microbiol Infect Dis. 2007;58:163–170. doi: 10.1016/j.diagmicrobio.2006.12.022. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

