BCDIN3D regulates tRNAHis 3' fragment processing
- PMID: 31329584
- PMCID: PMC6675128
- DOI: 10.1371/journal.pgen.1008273
BCDIN3D regulates tRNAHis 3' fragment processing
Abstract
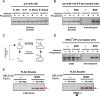
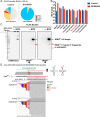
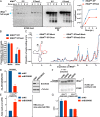
5' ends are important for determining the fate of RNA molecules. BCDIN3D is an RNA phospho-methyltransferase that methylates the 5' monophosphate of specific RNAs. In order to gain new insights into the molecular function of BCDIN3D, we performed an unbiased analysis of its interacting RNAs by Thermostable Group II Intron Reverse Transcriptase coupled to next generation sequencing (TGIRT-seq). Our analyses showed that BCDIN3D interacts with full-length phospho-methylated tRNAHis and miR-4454. Interestingly, we found that miR-4454 is not synthesized from its annotated genomic locus, which is a primer-binding site for an endogenous retrovirus, but rather by Dicer cleavage of mature tRNAHis. Sequence analysis revealed that miR-4454 is identical to the 3' end of tRNAHis. Moreover, we were able to generate this 'miRNA' in vitro through incubation of mature tRNAHis with Dicer. As found previously for several pre-miRNAs, a 5'P-tRNAHis appears to be a better substrate for Dicer cleavage than a phospho-methylated tRNAHis. Moreover, tRNAHis 3'-fragment/'miR-4454' levels increase in cells depleted for BCDIN3D. Altogether, our results show that in addition to microRNAs, BCDIN3D regulates tRNAHis 3'-fragment processing without negatively affecting tRNAHis's canonical function of aminoacylation.
Conflict of interest statement
I have read the journal's policy and the authors of this manuscript have the following competing interests: Thermostable Group II Intron Reverse Transcriptase (TGIRT) enzymes and methods for their use are the subject of patents and patent applications that have been licensed by the University of Texas and East Tennessee State University to InGex, LLC. AML and the University of Texas are minority equity holders in InGex, LLC, and AML and the University of Texas receive royalty payments from the sale of TGIRT enzymes and the licensing of intellectual property by InGex to other companies. The other authors declare no competing financial interests.
Figures

References
-
- Shatkin AJ. Capping of eucaryotic mRNAs. Cell. 1976;9(4 PT 2):645–53. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

