Unveil the transcriptional landscape at the Cryptococcus-host axis in mice and nonhuman primates
- PMID: 31329596
- PMCID: PMC6675133
- DOI: 10.1371/journal.pntd.0007566
Unveil the transcriptional landscape at the Cryptococcus-host axis in mice and nonhuman primates
Abstract
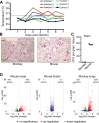
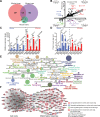
Pathogens and hosts require rapid modulation of virulence and defense mechanisms at the infection axis, but monitoring such modulations is challenging. In studying the human fungal pathogen Cryptococcus neoformans, mouse and rabbit infection models are often employed to shed light on the disease mechanisms but that may not be clinically relevant. In this study, we developed an animal infection model using the non-human primate cynomolgus monkey Macaca fascicularis. In addition, we systematically profiled and compared transcriptional responses between the infected mice and the cynomolgus monkey, using simultaneous or dual RNA next-generation sequencing. We demonstrated that there are shared but distinct transcriptional profiles between the two models following C. neoformans infection. Specifically, genes involved in immune and inflammatory responses are all upregulated. Osteoclastogenesis and insulin signaling are also significantly co-regulated in both models and disrupting an osteoclastogenesis-associated gene (OC-STAMP) or the insulin-signaling process significantly altered the host tolerance to C. neoformans. Moreover, C. neoformans was shown to activate metal sequestration, dampen the sugar metabolism, and control cell morphology during infection. Taking together, we described the development of a non-human primate model of cryptococcosis that allowed us to perform an in-depth analysis and comparison of transcriptome profiles during infections of two animal models and conceptually identify host genes important in disease responses. This study provides new insights in understanding fungal pathogenesis mechanisms that potentially facilitate the identification of novel drug targets for the treatment of cryptococcal infection.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

