Combining stable isotope analysis with DNA metabarcoding improves inferences of trophic ecology
- PMID: 31329604
- PMCID: PMC6645532
- DOI: 10.1371/journal.pone.0219070
Combining stable isotope analysis with DNA metabarcoding improves inferences of trophic ecology
Abstract
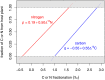
Knowing what animals eat is fundamental to our ability to understand and manage biodiversity and ecosystems, but researchers often must rely on indirect methods to infer trophic position and food intake. Using an approach that combines evidence from stable isotope analysis and DNA metabarcoding, we assessed the diet and trophic position of Anthene usamba butterflies, for which there are no known direct observations of larval feeding. An earlier study that analyzed adults rather than caterpillars of A. usamba inferred that this butterfly was aphytophagous, but we found that the larval guts of A. usamba and two known herbivorous lycaenid species contain chloroplast 16S sequences. Moreover, chloroplast barcoding revealed high sequence similarity between chloroplasts found in A. usamba guts and the chloroplasts of the Vachellia drepanolobium trees on which the caterpillars live. Stable isotope analysis provided further evidence that A. usamba caterpillars feed on V. drepanolobium, and the possibilities of strict herbivory versus limited omnivory in this species are discussed. These results highlight the importance of combining multiple approaches and considering ontogeny when using stable isotopes to infer trophic ecology where direct observations are difficult or impossible.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Garvey JE, Whiles M. Trophic Ecology. CRC Press; 2016.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

