Real-time PCR quantification of spliced X-box binding protein 1 (XBP1) using a universal primer method
- PMID: 31329612
- PMCID: PMC6645673
- DOI: 10.1371/journal.pone.0219978
Real-time PCR quantification of spliced X-box binding protein 1 (XBP1) using a universal primer method
Abstract
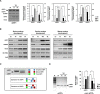
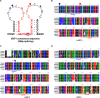
X-box binding protein 1 (XBP1) mRNA processing plays a crucial role in the unfolded protein response (UPR), which is activated in response to endoplasmic reticulum (ER) stress. Upon accumulation of the UPR-converted XBP1 mRNA splicing from an unspliced (u) XBP1 (inactive) isoform to the spliced (s) XBP1 (active) isoform, inositol-requiring enzyme 1 α (IRE1α) removes a 26-nucleotide intron from uXBP1 mRNA. Recent studies have reported the assessment of ER stress by examining the ratio of sXBP1 to uXBP1 mRNA (s/uXBP1 ratio) via densitometric analysis of PCR bands relative to increased levels of sXBP1 to uXBP1 using a housekeeping gene for normalization. However, this measurement is visualized by gel electrophoresis, making it very difficult to quantify differences between the two XBP1 bands and complicating data interpretation. Moreover, most commonly used housekeeping genes display an unacceptably high variable expression pattern of the s/uXBP1 ratio under different experimental conditions, such as various phases of development and different cell types, limiting their use as internal controls. For a more quantitative determination of XBP1 splicing activity, we measured the expression levels of total XBP1 (tXBP1: common region of s/uXBP1) and sXBP1 via real-time PCR using specific primer sets. We also designed universal real-time PCR primer sets capable of amplifying a portion of each u/s/tXBP1 mRNA that is highly conserved in eukaryotes, including humans, monkeys, cows, pigs, and mice. Therefore, we provide a more convenient and easily approachable quantitative real-time PCR method that can be used in various research fields to assess ER stress.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Bartoszewski R, Brewer JW, Rab A, Crossman DK, Bartoszewska S, Kapoor N, et al. The unfolded protein response (UPR)-activated transcription factor X-box-binding protein 1 (XBP1) induces microRNA-346 expression that targets the human antigen peptide transporter 1 (TAP1) mRNA and governs immune regulatory genes. J Biol Chem. 2011;286(48):41862–70. 10.1074/jbc.M111.304956 - DOI - PMC - PubMed
-
- Walter F, Schmid J, Dussmann H, Concannon CG, Prehn JH. Imaging of single cell responses to ER stress indicates that the relative dynamics of IRE1/XBP1 and PERK/ATF4 signalling rather than a switch between signalling branches determine cell survival. Cell Death Differ. 2015;22(9):1502–16. 10.1038/cdd.2014.241 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

