A Protocol for Extraction of Infective Viromes Suitable for Metagenomics Sequencing from Low Volume Fecal Samples
- PMID: 31330855
- PMCID: PMC6669555
- DOI: 10.3390/v11070667
A Protocol for Extraction of Infective Viromes Suitable for Metagenomics Sequencing from Low Volume Fecal Samples
Abstract
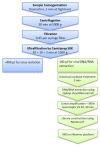
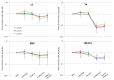
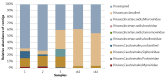
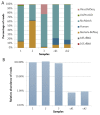
The human gut microbiome (GM) plays an important role in human health and diseases. However, while substantial progress has been made in understanding the role of bacterial inhabitants of the gut, much less is known regarding the viral component of the GM. Bacteriophages (phages) are viruses attacking specific host bacteria and likely play important roles in shaping the GM. Although metagenomic approaches have led to the discoveries of many new viruses, they remain largely uncultured as their hosts have not been identified, which hampers our understanding of their biological roles. Existing protocols for isolation of viromes generally require relatively high input volumes and are generally more focused on extracting nucleic acids of good quality and purity for down-stream analysis, and less on purifying viruses with infective capacity. In this study, we report the development of an efficient protocol requiring low sample input yielding purified viromes containing phages that are still infective, which also are of sufficient purity for genome sequencing. We validated the method through spiking known phages followed by plaque assays, qPCR, and metagenomic sequencing. The protocol should facilitate the process of culturing novel viruses from the gut as well as large scale studies on gut viromes.
Keywords: T4; c2; human gut phageome; human gut virome; isolation; microbiome; phage; phi29; phiX174; purification.
Conflict of interest statement
The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

