Stromal fibroblasts induce metastatic tumor cell clusters via epithelial-mesenchymal plasticity
- PMID: 31331982
- PMCID: PMC6653778
- DOI: 10.26508/lsa.201900425
Stromal fibroblasts induce metastatic tumor cell clusters via epithelial-mesenchymal plasticity
Abstract
Emerging evidence supports the hypothesis that multicellular tumor clusters invade and seed metastasis. However, whether tumor-associated stroma induces epithelial-mesenchymal plasticity in tumor cell clusters, to promote invasion and metastasis, remains unknown. We demonstrate herein that carcinoma-associated fibroblasts (CAFs) frequently present in tumor stroma drive the formation of tumor cell clusters composed of two distinct cancer cell populations, one in a highly epithelial (E-cadherinhiZEB1lo/neg: Ehi) state and another in a hybrid epithelial/mesenchymal (E-cadherinloZEB1hi: E/M) state. The Ehi cells highly express oncogenic cell-cell adhesion molecules, such as carcinoembryonic antigen-related cell adhesion molecule 5 (CEACAM5) and CEACAM6 that associate with E-cadherin, resulting in increased tumor cell cluster formation and metastatic seeding. The E/M cells also retain associations with Ehi cells, which follow the E/M cells leading to collective invasion. CAF-produced stromal cell-derived factor 1 and transforming growth factor-β confer the Ehi and E/M states as well as invasive and metastatic traits via Src activation in apposed human breast tumor cells. Taken together, these findings indicate that invasive and metastatic tumor cell clusters are induced by CAFs via epithelial-mesenchymal plasticity.
© 2019 Matsumura et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
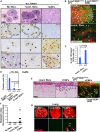


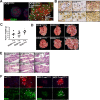
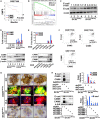







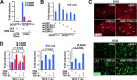





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous
