Environment Shapes the Accessible Daptomycin Resistance Mechanisms in Enterococcus faecium
- PMID: 31332078
- PMCID: PMC6761497
- DOI: 10.1128/AAC.00790-19
Environment Shapes the Accessible Daptomycin Resistance Mechanisms in Enterococcus faecium
Abstract
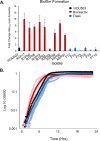
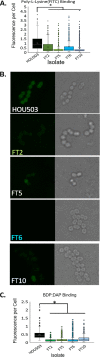
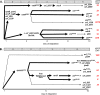
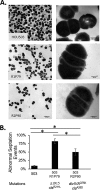
Daptomycin binds to bacterial cell membranes and disrupts essential cell envelope processes, leading to cell death. Bacteria respond to daptomycin by altering their cell envelopes to either decrease antibiotic binding to the membrane or by diverting binding away from septal targets. In Enterococcus faecalis, daptomycin resistance is typically coordinated by the three-component cell envelope stress response system, LiaFSR. Here, studying a clinical strain of multidrug-resistant Enterococcus faecium containing alleles associated with activation of the LiaFSR signaling pathway, we found that specific environments selected for different evolutionary trajectories, leading to high-level daptomycin resistance. Planktonic environments favored pathways that increased cell surface charge via yvcRS upregulation of dltABCD and mprF, causing a reduction in daptomycin binding. Alternatively, environments favoring complex structured communities, including biofilms, evolved both diversion and repulsion strategies via divIVA and oatA mutations, respectively. Both environments subsequently converged on cardiolipin synthase (cls) mutations, suggesting the importance of membrane modification across strategies. Our findings indicate that E. faecium can evolve diverse evolutionary trajectories to daptomycin resistance that are shaped by the environment to produce a combination of resistance strategies. The accessibility of multiple and different biochemical pathways simultaneously suggests that the outcome of daptomycin exposure results in a polymorphic population of resistant phenotypes, making E. faecium a recalcitrant nosocomial pathogen.
Keywords: Enterococcus; adaptive resistance; drug resistance evolution.
Copyright © 2019 American Society for Microbiology.
Figures

References
-
- CDC. 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, Atlanta, GA.
-
- Arias CA, Panesso D, McGrath DM, Qin X, Mojica MF, Miller C, Diaz L, Tran TT, Rincon S, Barbu EM, Reyes J, Roh JH, Lobos E, Sodergren E, Pasqualini R, Arap W, Quinn JP, Shamoo Y, Murray BE, Weinstock GM. 2011. Genetic basis for in vivo daptomycin resistance in enterococci. N Engl J Med 365:892–900. doi: 10.1056/NEJMoa1011138. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

