A Systems-level Characterization of the Differentiation of Human Embryonic Stem Cells into Mesenchymal Stem Cells
- PMID: 31332097
- PMCID: PMC6773553
- DOI: 10.1074/mcp.RA119.001356
A Systems-level Characterization of the Differentiation of Human Embryonic Stem Cells into Mesenchymal Stem Cells
Abstract
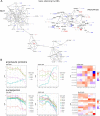
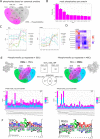
Mesenchymal stem/stromal cells (MSCs) are self-renewing multipotent cells with regenerative, secretory and immunomodulatory capabilities that are beneficial for the treatment of various diseases. To avoid the issues that come with using tissue-derived MSCs in therapy, MSCs may be generated by the differentiation of human embryonic stems cells (hESCs) in culture. However, the changes that occur during the differentiation process have not been comprehensively characterized. Here, we combined transcriptome, proteome and phosphoproteome profiling to perform an in-depth, multi-omics study of the hESCs-to-MSCs differentiation process. Based on RNA-to-protein correlation, we determined a set of high confidence genes that are important to differentiation. Among the earliest and strongest induced proteins with extensive differential phosphorylation was AHNAK, which we hypothesized to be a defining factor in MSC biology. We observed two distinct expression waves of developmental HOX genes and an AGO2-to-AGO3 switch in gene silencing. Exploring the kinetic of noncoding ORFs during differentiation, we mapped new functions to well annotated long noncoding RNAs (CARMN, MALAT, NEAT1, LINC00152) as well as new candidates which we identified to be important to the differentiation process. Phosphoproteome analysis revealed ESC and MSC-specific phosphorylation motifs with PAK2 and RAF1 as top predicted upstream kinases in MSCs. Our data represent a rich systems-level resource on ESC-to-MSC differentiation that will be useful for the study of stem cell biology.
Keywords: LC-MS/MS; RNA SEQ; cell differentiation; developmental biology; differentiation; gene expression; human embryonic stem cells; human mesenchymal stem cells; phosphoproteome; post-translational modifications; quantification; quantitative proteomics; stem cells; systems biology.
© 2019 Billing et al.
Conflict of interest statement
The authors declare no competing interests
Figures






References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

