Gram-scale total synthesis of teixobactin promoting binding mode study and discovery of more potent antibiotics
- PMID: 31332172
- PMCID: PMC6646333
- DOI: 10.1038/s41467-019-11211-y
Gram-scale total synthesis of teixobactin promoting binding mode study and discovery of more potent antibiotics
Abstract
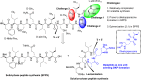
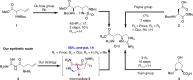
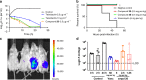
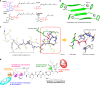
Teixobactin represents a new class of antibiotics with novel structure and excellent activity against Gram-positive pathogens and Mycobacterium tuberculosis. Herein, we report a one-pot reaction to conveniently construct the key building block L-allo-Enduracidine in 30-gram scale in just one hour and a convergent strategy (3 + 2 + 6) to accomplish a gram-scale total synthesis of teixobactin. Several analogs are described, with 20 and 26 identified as the most efficacious analogs with 3~8-fold and 2~4-fold greater potency against vancomycin resistant Enterococcus faecalis and methicillin-resistant Staphylococcus aureus respectively in comparison with teixobactin. In addition, they show high efficiency in Streptococcus pneumoniae septicemia mouse model and neutropenic mouse thigh infection model using methicillin-resistant Staphylococcus aureus. We also propose that the antiparallel β-sheet of teixobactin is important for its bioactivity and an antiparallel dimer of teixobactin is the minimal binding unit for lipid II via key amino acids variations and molecular docking.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

