Comparison of Four Complete Chloroplast Genomes of Medicinal and Ornamental Meconopsis Species: Genome Organization and Species Discrimination
- PMID: 31332227
- PMCID: PMC6646306
- DOI: 10.1038/s41598-019-47008-8
Comparison of Four Complete Chloroplast Genomes of Medicinal and Ornamental Meconopsis Species: Genome Organization and Species Discrimination
Erratum in
-
Author Correction: Comparison of Four Complete Chloroplast Genomes of Medicinal and Ornamental Meconopsis Species: Genome Organization and Species Discrimination.Sci Rep. 2019 Oct 17;9(1):15163. doi: 10.1038/s41598-019-51738-0. Sci Rep. 2019. PMID: 31619747 Free PMC article.
Abstract
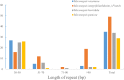
High-throughput sequencing of chloroplast genomes has been used to gain insight into the evolutionary relationships of plant species. In this study, we sequenced the complete chloroplast genomes of four species in the Meconopsis genus: M. racemosa, M. integrifolia (Maxim.) Franch, M. horridula and M. punicea. These plants grow in the wild and are recognized as having important medicinal and ornamental applications. The sequencing results showed that the size of the Meconopsis chloroplast genome ranges from 151864 to 153816 bp. A total of 127 genes comprising 90 protein-coding genes, 37 tRNA genes and 8 rRNA genes were observed in all four chloroplast genomes. Comparative analysis of the four chloroplast genomes revealed five hotspot regions (matK, rpoC2, petA, ndhF, and ycf1), which could potentially be used as unique molecular markers for species identification. In addition, the ycf1 gene may also be used as an effective molecular marker to distinguish Papaveraceae and determine the evolutionary relationships among plant species in the Papaveraceae family. Futhermore, these four genomes can provide valuable genetic information for other related studies.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Wang B, Song XH, Cheng CM. Advance in Ethnobotanical Investigation on Meconopsis. Chinese Academic Medical Magazine of Organisms. 2003;01:39–45.
-
- Guo Z, et al. Chemical constituents from a Tibetan medicine Meconopsis horridula. China Journal of Chinese Materia Medica. 2014;39:1152–1156. - PubMed
-
- Zhao Z, et al. Peng. Advances in studies on the classification, chemical composition and pharmacological action of Meconopsis as Tibetan medicines. China. Pharmacy. 2016;27:4391–4394.
-
- Chang Y, Wang XL, Tang XY, Yuan LY, Chen LH. A New Alkaloid from Meconopsis horridula. Natural Product Research and Development. 2017;29:731–734.
-
- Qu, Y. & Ou, Z. The research advancement on the genus Meconpsis. Northern Horticulture 191–194 (2012).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

