Virus Genotype-Dependent Transcriptional Alterations in Lipid Metabolism and Inflammation Pathways in the Hepatitis C Virus-infected Liver
- PMID: 31332246
- PMCID: PMC6646375
- DOI: 10.1038/s41598-019-46664-0
Virus Genotype-Dependent Transcriptional Alterations in Lipid Metabolism and Inflammation Pathways in the Hepatitis C Virus-infected Liver
Abstract
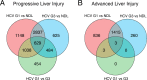
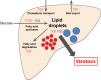
Despite advances in antiviral therapy, molecular drivers of Hepatitis C Virus (HCV)-related liver disease remain poorly characterised. Chronic infection with HCV genotypes (1 and 3) differ in presentation of liver steatosis and virological response to therapies, both to interferon and direct acting antivirals. To understand what drives these clinically important differences, liver expression profiles of patients with HCV Genotype 1 or 3 infection (n = 26 and 33), alcoholic liver disease (n = 8), and no liver disease (n = 10) were analysed using transcriptome-wide microarrays. In progressive liver disease, HCV genotype was the major contributor to altered liver gene expression with 2151 genes differentially expressed >1.5-fold between HCV Genotype 1 and 3. In contrast, only 6 genes were altered between the HCV genotypes in advanced liver disease. Induction of lipogenic, lipolytic, and interferon stimulated gene pathways were enriched in Genotype 1 injury whilst a broad range of immune-associated pathways were associated with Genotype 3 injury. The results are consistent with greater lipid turnover in HCV Genotype 1 patients. Moreover, the lower activity in inflammatory pathways associated with HCV genotype 1 is consistent with relative resistance to interferon-based therapy. This data provides a molecular framework to explain the clinical manifestations of HCV-associated liver disease.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

