Advancing the integration of multi-marker metabarcoding data in dietary analysis of trophic generalists
- PMID: 31332947
- PMCID: PMC6899665
- DOI: 10.1111/1755-0998.13060
Advancing the integration of multi-marker metabarcoding data in dietary analysis of trophic generalists
Abstract
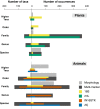
The application of DNA metabarcoding to dietary analysis of trophic generalists requires using multiple markers in order to overcome problems of primer specificity and bias. However, limited attention has been given to the integration of information from multiple markers, particularly when they partly overlap in the taxa amplified, and vary in taxonomic resolution and biases. Here, we test the use of a mix of universal and specific markers, provide criteria to integrate multi-marker metabarcoding data and a python script to implement such criteria and produce a single list of taxa ingested per sample. We then compare the results of dietary analysis based on morphological methods, single markers, and the proposed combination of multiple markers. The study was based on the analysis of 115 faeces from a small passerine, the Black Wheatears (Oenanthe leucura). Morphological analysis detected far fewer plant taxa (12) than either a universal 18S marker (57) or the plant trnL marker (124). This may partly reflect the detection of secondary ingestion by molecular methods. Morphological identification also detected far fewer taxa (23) than when using 18S (91) or the arthropod markers IN16STK (244) and ZBJ (231), though each method missed or underestimated some prey items. Integration of multi-marker data provided far more detailed dietary information than any single marker and estimated higher frequencies of occurrence of all taxa. Overall, our results show the value of integrating data from multiple, taxonomically overlapping markers in an example dietary data set.
Keywords: bird; diet; metabarcoding; morphological identification; overlapping markers; secondary predation.
© 2019 The Authors. Molecular Ecology Resources published by John Wiley & Sons Ltd.
Figures


References
-
- Aizpurua, O. , Budinski, I. , Georgiakakis, P. , Gopalakrishnan, S. , Ibañez, C. , Mata, V. , … Alberdi, A. (2018). Agriculture shapes the trophic niche of a bat preying on multiple pest arthropods across Europe: Evidence from DNA metabarcoding. Molecular Ecology, 27(3), 815–825. 10.1111/mec.14474 - DOI - PubMed
-
- Alberdi, A. , Aizpurua, O. , Gilbert, M. T. P. , & Bohmann, K. (2017). Scrutinizing key steps for reliable metabarcoding of environmental samples. Methods in Ecology and Evolution, 38(1), 42–49. 10.1111/2041-210X.12849 - DOI
-
- Barrett, R. T. , Camphuysen, K. , Anker‐Nilssen, T. , Chardine, J. W. , Furness, R. W. , Garthe, S. , … Veit, R. R. (2007). Diet studies of seabirds: A review and recommendations. ICES Journal of Marine Science, 64(9), 1675–1691. 10.1093/icesjms/fsm152 - DOI
-
- Barrientos, J. A. (Ed). (2004). Curso práctico de entomología. Barcelona, Spain: Asociación Española de Entomología, CIBIO‐Centro Iberoamericano de Biodiversidad & Universitat Autònoma de Barcelona.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

