PARK2 Mutation Causes Metabolic Disturbances and Impaired Survival of Human iPSC-Derived Neurons
- PMID: 31333417
- PMCID: PMC6624735
- DOI: 10.3389/fncel.2019.00297
PARK2 Mutation Causes Metabolic Disturbances and Impaired Survival of Human iPSC-Derived Neurons
Abstract
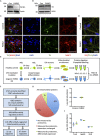
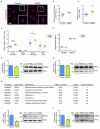
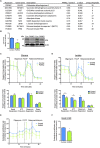
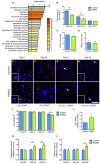
The protein parkin, encoded by the PARK2 gene, is vital for mitochondrial homeostasis, and although it has been implicated in Parkinson's disease (PD), the disease mechanisms remain unclear. We have applied mass spectrometry-based proteomics to investigate the effects of parkin dysfunction on the mitochondrial proteome in human isogenic induced pluripotent stem cell-derived neurons with and without PARK2 knockout (KO). The proteomic analysis quantified nearly 60% of all mitochondrial proteins, 119 of which were dysregulated in neurons with PARK2 KO. The protein changes indicated disturbances in oxidative stress defense, mitochondrial respiration and morphology, cell cycle control, and cell viability. Structural and functional analyses revealed an increase in mitochondrial area and the presence of elongated mitochondria as well as impaired glycolysis and lactate-supported respiration, leading to an impaired cell survival in PARK2 KO neurons. This adds valuable insight into the effect of parkin dysfunction in human neurons and provides knowledge of disease-related pathways that can potentially be targeted for therapeutic intervention.
Keywords: Parkinson’s; metabolism; mitochondria; oxidative stress; proteomics; survival.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

