Clear Victory for Chlamydia: The Subversion of Host Innate Immunity
- PMID: 31333596
- PMCID: PMC6619438
- DOI: 10.3389/fmicb.2019.01412
Clear Victory for Chlamydia: The Subversion of Host Innate Immunity
Abstract
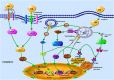
As obligate intracellular bacterial pathogens, members of the Chlamydia genera are the pivotal triggers for a wide range of infections, which can lead to blinding trachoma, pelvic inflammation, and respiratory diseases. Because of their restricted parasitism inside eukaryotic cells, the pathogens have to develop multiple strategies for adaptation with the hostile intracellular environment-intrinsically present in all host cells-to survive. The strategies that are brought into play at different stages of chlamydial development mainly involve interfering with diverse innate immune responses, such as innate immune recognition, inflammation, apoptosis, autophagy, as well as the manipulation of innate immune cells to serve as potential niches for chlamydial replication. This review will focus on the innate immune responses against chlamydial infection, highlighting the underlying molecular mechanisms used by the Chlamydia spp. to counteract host innate immune defenses. Insights into these subtle pathogenic mechanisms not only provide a rationale for the augmentation of immune responses against chlamydial infection but also open avenues for further investigation of the molecular mechanisms driving the survival of these clinically important pathogens in host innate immunity.
Keywords: Chlamydia; immune recognition; innate immune cells; innate immune response; survival and growth.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

