MITF Regulates Downstream Genes in Response to Vibrio parahaemolyticus Infection in the Clam Meretrix Petechialis
- PMID: 31333673
- PMCID: PMC6620822
- DOI: 10.3389/fimmu.2019.01547
MITF Regulates Downstream Genes in Response to Vibrio parahaemolyticus Infection in the Clam Meretrix Petechialis
Abstract
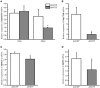
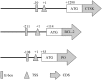
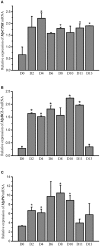
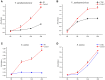
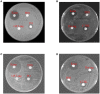
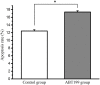
The microphthalmia-associated transcription factor (MITF) is a basic helix-loop-helix-leucine zipper protein that plays a key role in cell proliferation, survival and immune defense through the direct transcriptional control of downstream genes. We have found that MITF participates in the immune response to Vibrio parahaemolyticus infection in the clam Meretrix petechialis. In this study, we focused on how MITF functions in immunity. First, PO, CTSK, and BCL-2 were identified as the target genes of MpMITF in the clam by RNAi. EMSAs showed direct binding between the MpMITF protein and the E-box of the MpPO, MpCTSK, and MpBCL-2 promoters. Yeast one-hybrid assays also suggested that MpMITF could activate the expression of these three downstream genes. These results demonstrated that the transcriptional expression of MpPO, MpCTSK, and MpBCL-2 is directly regulated by MpMITF. Second, we analyzed the roles of MpPO, MpCTSK, and MpBCL-2 in clam immunity. The mRNA expression of MpPO, MpCTSK, and MpBCL-2 increased significantly after V. parahaemolyticus challenge, which implied that these genes might take part in the immune defense against V. parahaemolyticus challenge in clams. The purified recombinant proteins, MpPO and MpCTSK, inhibited the growth of V. parahaemolyticus. Additionally, the apoptosis rate of clam haemocytes rose significantly when the activity of MpBCL-2 was suppressed. These results revealed that MpPO, MpCTSK, and MpBCL-2 are involved in the immune defense against V. parahaemolyticus. This study supports the idea that the MpMITF pathway plays a key role in immune defense through the direct regulation of the downstream genes MpPO, MpCTSK, and MpBCL-2 in the clam, M. petechialis.
Keywords: MITF; Vibrio parahaemolyticus; clam; immune response; signaling pathway.
Figures


References
-
- Hughes MJ, Lingrel JB, Krakowskyt JM, Anderson KP. A helix-loop-helix transcription factor-like gene is located at the mi locus. J Biol Chem. (1993) 268:20687–90. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

