ZC4H2 stabilizes RNF220 to pattern ventral spinal cord through modulating Shh/Gli signaling
- PMID: 31336385
- PMCID: PMC7288745
- DOI: 10.1093/jmcb/mjz087
ZC4H2 stabilizes RNF220 to pattern ventral spinal cord through modulating Shh/Gli signaling
Abstract
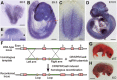
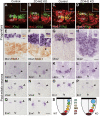
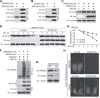
ZC4H2 encodes a C4H2 type zinc-finger nuclear factor, the mutation of which has been associated with disorders with various clinical phenotypes in human, including developmental delay, intellectual disability and dystonia. ZC4H2 has been suggested to regulate spinal cord patterning in zebrafish as a co-factor for RNF220, an ubiquitin E3 ligase involved in Gli signaling. Here we showed that ZC4H2 and RNF220 knockout animals phenocopy each other in spinal patterning in both mouse and zebrafish, with mispatterned progenitor and neuronal domains in the ventral spinal cord. We showed evidence that ZC4H2 is required for the stability of RNF220 and also proper Gli ubiquitination and signaling in vivo. Our data provides new insights into the possible etiology of the neurodevelopmental impairments observed in ZC4H2-associated syndromes.
Keywords: Gli signaling; RNF220; ZC4H2; patterning; spinal cord.
© The Author(s) (2019). Published by Oxford University Press on behalf of Journal of Molecular Cell Biology, IBCB, SIBS, CAS.
Figures



References
-
- Briscoe J., Pierani A., Jessell T.M., et al. (2000). A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell 101, 435–445. - PubMed
-
- Cheng X.N., Shao M., and Shi D.L. (2018). Mutation of Frizzled8a delays neural retinal cell differentiation and results in microphthalmia in zebrafish. Int. J. Dev. Biol. 62, 285–291. - PubMed
-
- Dessaud E., McMahon A.P., and Briscoe J. (2008). Pattern formation in the vertebrate neural tube: a sonic Hedgehog morphogen-regulated transcriptional network. Development 135, 2489–2503. - PubMed
-
- Jessell T.M. (2000). Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 1, 20–29. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

