Statistical Workflow for Feature Selection in Human Metabolomics Data
- PMID: 31336989
- PMCID: PMC6680705
- DOI: 10.3390/metabo9070143
Statistical Workflow for Feature Selection in Human Metabolomics Data
Abstract
High-throughput metabolomics investigations, when conducted in large human cohorts, represent a potentially powerful tool for elucidating the biochemical diversity underlying human health and disease. Large-scale metabolomics data sources, generated using either targeted or nontargeted platforms, are becoming more common. Appropriate statistical analysis of these complex high-dimensional data will be critical for extracting meaningful results from such large-scale human metabolomics studies. Therefore, we consider the statistical analytical approaches that have been employed in prior human metabolomics studies. Based on the lessons learned and collective experience to date in the field, we offer a step-by-step framework for pursuing statistical analyses of cohort-based human metabolomics data, with a focus on feature selection. We discuss the range of options and approaches that may be employed at each stage of data management, analysis, and interpretation and offer guidance on the analytical decisions that need to be considered over the course of implementing a data analysis workflow. Certain pervasive analytical challenges facing the field warrant ongoing focused research. Addressing these challenges, particularly those related to analyzing human metabolomics data, will allow for more standardization of as well as advances in how research in the field is practiced. In turn, such major analytical advances will lead to substantial improvements in the overall contributions of human metabolomics investigations.
Keywords: high-dimensional data; large-scale metabolomics; statistical methods.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

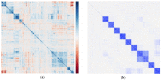
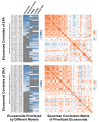
References
-
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. - DOI
-
- Mayers J.R., Wu C., Clish C.B., Kraft P., Torrence M.E., Fiske B.P., Yuan C., Bao Y., Townsend M.K., Tworoger S.S. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat. Med. 2014;20:1193–1198. doi: 10.1038/nm.3686. - DOI - PMC - PubMed

