Plant Phytochromes and their Phosphorylation
- PMID: 31337079
- PMCID: PMC6678601
- DOI: 10.3390/ijms20143450
Plant Phytochromes and their Phosphorylation
Abstract
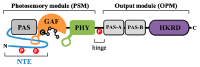
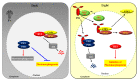
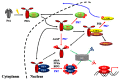
Extensive research over several decades in plant light signaling mediated by photoreceptors has identified the molecular mechanisms for how phytochromes regulate photomorphogenic development, which includes degradation of phytochrome-interacting factors (PIFs) and inactivation of COP1-SPA complexes with the accumulation of master transcription factors for photomorphogenesis, such as HY5. However, the initial biochemical mechanism for the function of phytochromes has not been fully elucidated. Plant phytochromes have long been known as phosphoproteins, and a few protein phosphatases that directly interact with and dephosphorylate phytochromes have been identified. However, there is no report thus far of a protein kinase that acts on phytochromes. On the other hand, plant phytochromes have been suggested as autophosphorylating serine/threonine protein kinases, proposing that the kinase activity might be important for their functions. Indeed, the autophosphorylation of phytochromes has been reported to play an important role in the regulation of plant light signaling. More recently, evidence that phytochromes function as protein kinases in plant light signaling has been provided using phytochrome mutants displaying reduced kinase activities. In this review, we highlight recent advances in the reversible phosphorylation of phytochromes and their functions as protein kinases in plant light signaling.
Keywords: autophosphorylation; light signaling; plant photoreceptors; protein kinase; reversible phosphorylation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Regulation of Plant Photoresponses by Protein Kinase Activity of Phytochrome A.Int J Mol Sci. 2023 Jan 20;24(3):2110. doi: 10.3390/ijms24032110. Int J Mol Sci. 2023. PMID: 36768431 Free PMC article. Review.
-
Phytochromes and Phytochrome Interacting Factors.Plant Physiol. 2018 Feb;176(2):1025-1038. doi: 10.1104/pp.17.01384. Epub 2017 Nov 14. Plant Physiol. 2018. PMID: 29138351 Free PMC article. Review.
-
Phytochrome phosphorylation in plant light signaling.Front Plant Sci. 2024 Mar 12;15:1259720. doi: 10.3389/fpls.2024.1259720. eCollection 2024. Front Plant Sci. 2024. PMID: 38545394 Free PMC article. Review.
-
Multiple kinases promote light-induced degradation of PIF1.Plant Signal Behav. 2011 Aug;6(8):1119-21. doi: 10.4161/psb.6.8.16049. Epub 2011 Aug 1. Plant Signal Behav. 2011. PMID: 21758014 Free PMC article. Review.
-
Decoding of light signals by plant phytochromes and their interacting proteins.Annu Rev Plant Biol. 2008;59:281-311. doi: 10.1146/annurev.arplant.59.032607.092859. Annu Rev Plant Biol. 2008. PMID: 18257712 Review.
Cited by
-
Molecular Evolution and Interaction of Membrane Transport and Photoreception in Plants.Front Genet. 2019 Oct 11;10:956. doi: 10.3389/fgene.2019.00956. eCollection 2019. Front Genet. 2019. PMID: 31681411 Free PMC article. Review.
-
Phytochrome A in plants comprises two structurally and functionally distinct populations - water-soluble phyA' and amphiphilic phyA″.Biophys Rev. 2022 Jul 1;14(4):905-921. doi: 10.1007/s12551-022-00974-2. eCollection 2022 Aug. Biophys Rev. 2022. PMID: 36124260 Free PMC article. Review.
-
Regulation of Photomorphogenic Development by Plant Phytochromes.Int J Mol Sci. 2019 Dec 6;20(24):6165. doi: 10.3390/ijms20246165. Int J Mol Sci. 2019. PMID: 31817722 Free PMC article. Review.
-
Light quality, oxygenic photosynthesis and more.Photosynthetica. 2022 Jan 6;60(1):25-28. doi: 10.32615/ps.2021.055. eCollection 2022. Photosynthetica. 2022. PMID: 39648998 Free PMC article. Review.
-
Advanced genes expression pattern greatly contributes to divergence in Verticillium wilt resistance between Gossypium barbadense and Gossupium hirsutum.Front Plant Sci. 2022 Aug 1;13:979585. doi: 10.3389/fpls.2022.979585. eCollection 2022. Front Plant Sci. 2022. PMID: 35979082 Free PMC article.
References
-
- Kami C., Lorrain S., Hornitschek P., Fankhauser C. Light-regulated plant growth and development. Curr. Top. Dev. Biol. 2010;91:29–66. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

